Abstract
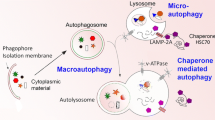
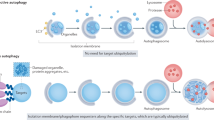
The autophagy–lysosome pathway is a highly conserved bulk degradation system in eukaryotes. During starvation, cytoplasmic constituents are non-selectively degraded by autophagy, and the resulting amino acids are utilized for cell survival. By taking advantage of mouse genetics, many physiological functions of mammalian autophagy have been uncovered. Growing lines of evidences have revealed the essential role of constitutive (or basal) autophagy in cellular homeostasis through its selectivity. p62, one of the selective substrates for autophagy, plays a key role in the formation of cytoplasmic proteinaceous inclusion, a hallmark of conformational diseases such as Alzheimer’s disease, Parkinson’s disease, and various chronic liver disorders. In this review, we discuss the physiological roles of the selective turnover of p62 by autophagy and their molecular mechanisms.




Similar content being viewed by others
Abbreviations
- Ape1:
-
Aminopeptidase I
- Atg:
-
Autophagy-related
- DISC:
-
Death-inducing signaling complex
- LC3:
-
Microtubule-associated protein 1 light chain 3
- LIR:
-
LC3-interacting region
- LRS:
-
LC3 recognition sequence
- PB1:
-
Phox and Bem1p
- PE:
-
Phosphatidylethanolamine
- UBA:
-
Ubiquitin-associated domain
- UPS:
-
Ubiquitin–proteasome system
References
Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426:895–899
Mizushima N (2007) Autophagy: process and function. Genes Dev 21:2861–2873
Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10:458–467
Yang Z, Klionsky DJ (2009) An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 335:1–32
Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333:169–174
Kuma A, Hatano M, Matsui M et al (2004) The role of autophagy during the early neonatal starvation period. Nature 432:1032–1036
Tsukamoto S, Kuma A, Murakami M et al (2008) Autophagy is essential for preimplantation development of mouse embryos. Science 321:117–120
Komatsu M, Ueno T, Waguri S et al (2007) Constitutive autophagy: vital role in clearance of unfavorable proteins in neurons. Cell Death Differ 14:887–894
Komatsu M, Waguri S, Koike M et al (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131:1149–1163
Komatsu M, Waguri S, Ueno T et al (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169:425–434
Komatsu M, Waguri S, Chiba T et al (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884
Hara T, Nakamura K, Matsui M et al (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889
Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130:165–178
Kabeya Y, Mizushima N, Ueno T et al (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728
Fujita N, Itoh T, Omori H et al (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19:2092–2100
Bjorkoy G, Lamark T, Brech A et al (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614
Pankiv S, Clausen TH, Lamark T et al (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145
Laurin N, Brown JP, Morissette J, Raymond V (2002) Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in Paget disease of bone. Am J Hum Genet 70:1582–1588
Moscat J, Diaz-Meco MT, Albert A, Campuzano S (2006) Cell signaling and function organized by PB1 domain interactions. Mol Cell 23:631–640
Jin Z, Li Y, Pitti R et al (2009) Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137:721–735
Nakaso K, Yoshimoto Y, Nakano T et al (2004) Transcriptional activation of p62/A170/ZIP during the formation of the aggregates: possible mechanisms and the role in Lewy body formation in Parkinson’s disease. Brain Res 1012:42–51
Nagaoka U, Kim K, Jana NR et al (2004) Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J Neurochem 91:57–68
Nan L, Wu Y, Bardag-Gorce F et al (2004) p62 is involved in the mechanism of Mallory body formation. Exp Mol Pathol 77:168–175
Ichimura Y, Kumanomidou T, Sou YS et al (2008) Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem 283:22847–22857
Shvets E, Fass E, Scherz-Shouval R, Elazar Z (2008) The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J Cell Sci 121:2685–2695
Noda NN, Kumeta H, Nakatogawa H et al (2008) Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13:1211–1218
Schweers RL, Zhang J, Randall MS et al (2007) NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA 104:19500–19505
Sandoval H, Thiagarajan P, Dasgupta SK et al (2008) Essential role for Nix in autophagic maturation of erythroid cells. Nature 454:232–235
Novak I, Kirkin V, McEwan DG et al (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 11:45–51
Satoo K, Noda NN, Kumeta H et al (2009) The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J 28:1341–1350
Narendra DP, Jin SM, Tanaka A et al (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8:e1000298
Narendra D, Tanaka A, Suen DF, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183:795–803
Geisler S, Holmstrom KM, Skujat D et al (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12:119–131
Duran A, Linares JF, Galvez AS et al (2008) The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell 13:343–354
Komatsu M, Kurokawa H, Waguri S et al (2010) The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12:213–223
Itoh K, Chiba T, Takahashi S et al (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322
Nakai A, Yamaguchi O, Takeda T et al (2007) The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med 13:619–624
Ebato C, Uchida T, Arakawa M et al (2008) Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab 8:325–332
Kirkin V, Lamark T, Sou YS et al (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33:505–516
Acknowledgments
This work is supported by Grants-in-Aid for Science Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (M.K.) and from the Japan Science and Technology Agency and the Ministry of Education, Science and Culture of Japan (M.K.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ichimura, Y., Komatsu, M. Selective degradation of p62 by autophagy. Semin Immunopathol 32, 431–436 (2010). https://doi.org/10.1007/s00281-010-0220-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-010-0220-1