Abstract
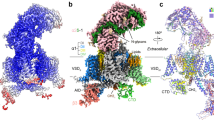
The voltage-gated calcium (Cav) channels convert membrane electrical signals to intracellular Ca2+-mediated events. Among the ten subtypes of Cav channel in mammals, Cav1.1 is specified for the excitation–contraction coupling of skeletal muscles. Here we present the cryo-electron microscopy structure of the rabbit Cav1.1 complex at a nominal resolution of 3.6 Å. The inner gate of the ion-conducting α1-subunit is closed and all four voltage-sensing domains adopt an ‘up’ conformation, suggesting a potentially inactivated state. The extended extracellular loops of the pore domain, which are stabilized by multiple disulfide bonds, form a windowed dome above the selectivity filter. One side of the dome provides the docking site for the α2δ-1-subunit, while the other side may attract cations through its negative surface potential. The intracellular I–II and III–IV linker helices interact with the β1a-subunit and the carboxy-terminal domain of α1, respectively. Classification of the particles yielded two additional reconstructions that reveal pronounced displacement of β1a and adjacent elements in α1. The atomic model of the Cav1.1 complex establishes a foundation for mechanistic understanding of excitation–contraction coupling and provides a three-dimensional template for molecular interpretations of the functions and disease mechanisms of Cav and Nav channels.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Data deposits
The atomic coordinates of the overall structure of the rabbit Cav1.1 complex shown in Fig. 1 have been deposited in the Protein Data Bank (PDB) under accession number 5GJV, and those of the structure built on the class II reconstruction have been deposited in the PDB under accession number 5GJW. The cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-9513 (class I), EMD-9514 (class Ia), and EMD-9515 (class II).
References
Clapham, D. E. Calcium signaling. Cell 131, 1047–1058 (2007)
Zamponi, G. W., Striessnig, J., Koschak, A. & Dolphin, A. C. The physiology, pathology, and pharmacology of voltage-gated calcium channels and their future therapeutic potential. Pharmacol. Rev. 67, 821–870 (2015)
Catterall, W. A. Voltage-gated calcium channels. Cold Spring Harb. Perspect. Biol. 3, a003947 (2011)
Nowycky, M. C., Fox, A. P. & Tsien, R. W. Three types of neuronal calcium channel with different calcium agonist sensitivity. Nature 316, 440–443 (1985)
Ertel, E. A. et al. Nomenclature of voltage-gated calcium channels. Neuron 25, 533–535 (2000)
Hille, B. Ion Channels of Excitable Membranes, 814 (Sinauer, 2001)
Kang, M. G. & Campbell, K. P. γ Subunit of voltage-activated calcium channels. J. Biol. Chem. 278, 21315–21318 (2003)
Davies, A. et al. Functional biology of the α2δ subunits of voltage-gated calcium channels. Trends Pharmacol. Sci. 28, 220–228 (2007)
Buraei, Z. & Yang, J. The β-subunit of voltage-gated Ca2+ channels. Physiol. Rev. 90, 1461–1506 (2010)
Kohlhardt, M. & Fleckenstein, A. Inhibition of the slow inward current by nifedipine in mammalian ventricular myocardium. Naunyn Schmiedebergs Arch. Pharmacol. 298, 267–272 (1977)
Fleckenstein, A. History of calcium antagonists. Circ. Res. 52, I3–I16 (1983)
Tanabe, T. et al. Primary structure of the receptor for calcium channel blockers from skeletal muscle. Nature 328, 313–318 (1987)
Bannister, R. A. & Beam, K. G. Ca(V)1.1: The atypical prototypical voltage-gated Ca2+ channel. Biochim. Biophys. Acta 1828, 1587–1597 (2013)
Rios, E. & Brum, G. Involvement of dihydropyridine receptors in excitation–contraction coupling in skeletal muscle. Nature 325, 717–720 (1987)
Tanabe, T., Beam, K. G., Powell, J. A. & Numa, S. Restoration of excitation–contraction coupling and slow calcium current in dysgenic muscle by dihydropyridine receptor complementary DNA. Nature 336, 134–139 (1988)
Block, B. A., Imagawa, T., Campbell, K. P. & Franzini-Armstrong, C. Structural evidence for direct interaction between the molecular components of the transverse tubule/sarcoplasmic reticulum junction in skeletal muscle. J. Cell Biol. 107, 2587–2600 (1988)
Adams, B. A., Tanabe, T., Mikami, A., Numa, S. & Beam, K. G. Intramembrane charge movement restored in dysgenic skeletal muscle by injection of dihydropyridine receptor cDNAs. Nature 346, 569–572 (1990)
Takekura, H., Bennett, L., Tanabe, T., Beam, K. G. & Franzini-Armstrong, C. Restoration of junctional tetrads in dysgenic myotubes by dihydropyridine receptor cDNA. Biophys. J. 67, 793–803 (1994)
Wu, J. et al. Structure of the voltage-gated calcium channel Cav1.1 complex. Science 350, aad2395 (2015)
Simske, J. S. Claudins reign: the claudin/EMP/PMP22/γ channel protein family in C. elegans. Tissue Barriers 1, e25502 (2013)
Davies, A. et al. The α2δ-subunits of voltage-gated calcium channels form GPI-anchored proteins, a posttranslational modification essential for function. Proc. Natl Acad. Sci. USA 107, 1654–1659 (2010)
Cantí, C. et al. The metal-ion-dependent adhesion site in the Von Willebrand factor-A domain of α2δ-subunits is key to trafficking voltage-gated Ca2+ channels. Proc. Natl Acad. Sci. USA 102, 11230–11235 (2005)
Yang, J., Ellinor, P. T., Sather, W. A., Zhang, J. F. & Tsien, R. W. Molecular determinants of Ca2+ selectivity and ion permeation in L-type Ca2+ channels. Nature 366, 158–161 (1993)
Ellinor, P. T., Yang, J., Sather, W. A., Zhang, J. F. & Tsien, R. W. Ca2+ channel selectivity at a single locus for high-affinity Ca2+ interactions. Neuron 15, 1121–1132 (1995)
Cloues, R. K., Cibulsky, S. M. & Sather, W. A. Ion interactions in the high-affinity binding locus of a voltage-gated Ca2+ channel. J. Gen. Physiol. 116, 569–586 (2000)
Tang, L. et al. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature 505, 56–61 (2014)
Payandeh, J., Scheuer, T., Zheng, N. & Catterall, W. A. The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011)
Zhang, X. et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 486, 130–134 (2012)
Ahuja, S. et al. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science 350, aac5464 (2015)
Armstrong, C. M. & Bezanilla, F. Currents related to movement of the gating particles of the sodium channels. Nature 242, 459–461 (1973)
Noda, M. et al. Primary structure of Electrophorus electricus sodium channel deduced from cDNA sequence. Nature 312, 121–127 (1984)
Tao, X., Lee, A., Limapichat, W., Dougherty, D. A. & MacKinnon, R. A gating charge transfer center in voltage sensors. Science 328, 67–73 (2010)
Andronache, Z. et al. The auxiliary-subunit γ1 of the skeletal muscle L-type Ca2+ channel is an endogenous Ca2+ antagonist. Proc. Natl Acad. Sci. USA 104, 17885–17890 (2007)
Almagor, L. et al. The role of a voltage-dependent Ca2+ channel intracellular linker: a structure-function analysis. J. Neurosci. 32, 7602–7613 (2012)
Yan, Z. et al. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature 517, 50–55 (2015)
Zalk, R. et al. Structure of a mammalian ryanodine receptor. Nature 517, 44–49 (2015)
Efremov, R. G., Leitner, A., Aebersold, R. & Raunser, S. Architecture and conformational switch mechanism of the ryanodine receptor. Nature 517, 39–43 (2015)
Gregg, R. G. et al. Absence of the β subunit (cchb1) of the skeletal muscle dihydropyridine receptor alters expression of the α1 subunit and eliminates excitation-contraction coupling. Proc. Natl Acad. Sci. USA 93, 13961–13966 (1996)
Cheng, W., Altafaj, X., Ronjat, M. & Coronado, R. Interaction between the dihydropyridine receptor Ca2+ channel β-subunit and ryanodine receptor type 1 strengthens excitation-contraction coupling. Proc. Natl Acad. Sci. USA 102, 19225–19230 (2005)
Schredelseker, J. et al. The β1a subunit is essential for the assembly of dihydropyridine-receptor arrays in skeletal muscle. Proc. Natl Acad. Sci. USA 102, 17219–17224 (2005)
Stotz, S. C., Jarvis, S. E. & Zamponi, G. W. Functional roles of cytoplasmic loops and pore lining transmembrane helices in the voltage-dependent inactivation of HVA calcium channels. J. Physiol. (Lond.) 554, 263–273 (2004)
Wang, C., Chung, B. C., Yan, H., Lee, S. Y. & Pitt, G. S. Crystal structure of the ternary complex of a NaV C-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Structure 20, 1167–1176 (2012)
Wang, C. et al. Structural analyses of Ca2+/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation. Nature Commun. 5, 4896 (2014)
Lee, K. S., Marban, E. & Tsien, R. W. Inactivation of calcium channels in mammalian heart cells: joint dependence on membrane potential and intracellular calcium. J. Physiol. (Lond.) 364, 395–411 (1985)
Zühlke, R. D., Pitt, G. S., Deisseroth, K., Tsien, R. W. & Reuter, H. Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature 399, 159–162 (1999)
Armstrong, C.M. & Bezanilla, F. Inactivation of the sodium channel. II. Gating current experiments. J Gen. Physiol. 70, 567–90 (1977)
Rohl, C.A. et al. Solution structure of the sodium channel inactivation gate. Biochemistry 38, 855–61 (1999)
DeLano, W. L. The PyMOL Molecular Graphics System (2002)
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996)
Pettersen, E. F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Yuan, Z. F. et al. pParse: a method for accurate determination of monoisotopic peaks in high-resolution mass spectra. Proteomics 12, 226–235 (2012)
Lu, S. et al. Mapping native disulfide bonds at a proteome scale. Nature Methods 12, 329–331 (2015)
Li, X., Zheng, S., Agard, D. A. & Cheng, Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J. Struct. Biol. 192, 174–178 (2015)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods 10, 584–590 (2013)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Gong, X. et al. Structural insights into the Niemann-Pick C1 (NPC1)-mediated cholesterol transfer and Ebola infection. Cell 165, 1467–1478 (2016)
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nature Methods 11, 63–65 (2014)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
DiMaio, F., Tyka, M. D., Baker, M. L., Chiu, W. & Baker, D. Refinement of protein structures into low-resolution density maps using Rosetta. J. Mol. Biol. 392, 181–190 (2009)
Song, Y. et al. High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013)
DiMaio, F. et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nature Methods 12, 361–365 (2015)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Nicholls, R. A., Fischer, M., McNicholas, S. & Murshudov, G. N. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr. D 70, 2487–2499 (2014)
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal-subunit. Science 343, 1485–1489 (2014)
Guo, J. et al. Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana. Nature 531, 196–201 (2016)
Kintzer, A. F. & Stroud, R. M. Structure, inhibition and regulation of two-pore channel TPC1 from Arabidopsis thaliana. Nature 531, 258–264 (2016)
Acknowledgements
We thank J. Lei, Y. Xu, and X. Li for technical support. We thank the Tsinghua University Branch of the China National Center for Protein Sciences (Beijing) for providing the facility support. The computation was completed on the ‘Explorer 100’ cluster system of Tsinghua National Laboratory for Information Science and Technology. This work was supported by funds from the Ministry of Science and Technology of China (2015CB9101012014, 2016YFA0500402, ZX09507003006) and the National Natural Science Foundation of China (project 31321062). The research of N.Y. was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute and an endowed professorship from Bayer Healthcare.
Author information
Authors and Affiliations
Contributions
N.Y. conceived the project. N.Y., J.W., Z.Y., and Z.L. designed all experiments. J.W., Z.Y., Z.L., X.Q., and S.L. performed experiments. J.W., Z.Y., Z.L., and Q.Z. conducted the cryo-EM analysis. All authors contributed to data analysis. J.W., M.D., and Q.Z. contributed to manuscript preparation. N.Y. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks W. Sather, G. Zamponi and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Cryo-EM analysis of the rabbit Cav1.1 complex.
a, A representative electron micrograph of the Cav1.1 complex. Scale bar, 400 Å. b, Two-dimensional class averages of the electron micrographs. The box size and circle diameter are 264 Å and 220 Å, respectively. c, Angular distribution for the final reconstruction of the Cav1.1 complex. Each cylinder represents one view and the height of the cylinder is proportional to the number of particles for that view. d, The gold-standard Fourier shell correlation curves for the EM maps. See Extended Data Fig. 2 and Methods for details of the three classes. e, Fourier shell correlation curves of the refined model versus the overall 3.6 Å map that it was refined against (black); of the model refined in the first of the two independent maps used for the gold-standard Fourier shell correlation curves versus that same map (red); and of the model refined in the first of the two independent maps versus the second independent map (green). The small difference between the red and green curves indicates that the refinement of the atomic coordinates did not suffer from overfitting. f, The overall EM maps of the Cav1.1 complex are colour-coded to indicate the range of resolutions. See Extended Data Fig. 2 for the definition of classes I, Ia, and II. The resolution maps are calculated with ResMap60.
Extended Data Figure 2 Flowchart for cryo-EM data processing of the Cav1.1 complex.
See ‘Image processing’ in Methods for details of (a) the flowchart and (b) the random-phase 3D classification method58.
Extended Data Figure 3 The new map reconstructed at 3.6 Å exhibits qualitative improvement over the reported 4.2 Å map.
The same four perpendicular side views are shown for the published 4.2 Å map (a), the new 3.6 Å map presented here (b), and their superimposition (c). The EM maps were generated in Chimera.
Extended Data Figure 4 Topology and EM maps of the α2δ-1-subunit.
a, Topology of the α2δ-subunit. The domains are coloured the same as the domain structures shown in Fig. 2. b, EM map for the overall α2δ-1-subunit. The EM map for the C-terminal stretch of the δ-subunit is shown at the bottom and coloured orange. As seen in the EM map, the consecutive density of the δ-subunit extends slightly beyond Cys1074. The additional density would correspond to Gly1075 if the C terminus were not cleaved during maturation, or alternatively, the ethanolamine of glycophosphatidylinositol (GPI) that modifies Cys1074. No peptide was detected for sequences after Cys1074 in the MS analysis of the purified Cav1.1 complex, and no additional density was found that may correspond to the C-terminal sequences of the δ-subunit. We thereby assigned an ethanolamine to the density following Cys1074. c, EM maps of representative β-strands in the α2-subunit. Left: the EM maps for the β-sheets in the VWA (upper) and cache1 (bottom) domains. Right: the EM maps for representative β-strands in the α2-subunit. NAG, N-acetylglucosamine. d, EM maps of representative α-helices in the α2-subunit. e, The EM maps for the MIDAS motif in the VWA domain and the loop between S1 and S2 in the first VSD of α1 (designated the L1–2I loop). The density corresponding to the cation is coloured magenta. The maps were generated using class I reconstruction and contoured at 6–8σ in PyMol.
Extended Data Figure 5 Representative EM maps for segments in the α1-subunit.
The EM maps for the S2 and S4 segments in the four VSDs (a), the S5 (b), and S6 (c) segments in the pore domain are shown. d, The EM map of the selectivity filter and the supporting P1 and P2 helices. Side-chain assignment was assisted by bulky residues in P1 and P2 helices as exemplified in the right panel. A consecutive stretch of density was observed along the selectivity filter vestibule. e, The densities that may correspond to lipid molecules bound to the α1-subunit. The maps were made using class I reconstruction and contoured at 4–6σ in PyMol.
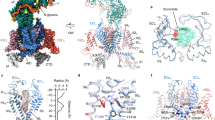
Extended Data Figure 6 Structural features of the pore domain of the α1-subunit.
a, The structure of the pore domain in four perpendicular side views. The disulfide bonds and glycosyl moieties are shown as sticks. Left: the densities below selectivity filter that may correspond to lipid tails are shown at 5σ. The modelled lipids are shown as yellow sticks. Similar densities that penetrate the side portals of the central cavity of the pore domain were previously observed in the structures of NavAb27 and NavRh28. b, The negative surface potential of the extracellular loops and the fenestrations of the pore domain. The surface electrostatic potential was calculated in PyMol. The fenestrations in the transmembrane region are highlighted by orange circles. c, The potential extracellular Ca2+ entrances through the windowed dome of the α1-subunit. Two potential entrances for Ca2+ are contoured with yellow lines in the top panels. The residues that underlie the negative surface potentials are shown in the bottom panels. d, The interface between the α2δ-1- and α1-subunits. The L1–2I, L5II, and L5III loops form the docking site for the VWA and cache1 domains of the α2δ-1-subunit.
Extended Data Figure 7 Putative Ca2+ coordination in the selectivity filter vestibule.
a–c, The densities that may correspond to bound Ca2+ ions in the selectivity filter vestibule exhibit distinct features in the present and previously published maps19. The Ca2+ concentrations for the samples that yielded the new 3.6 Å and previous 4.2 Å maps are 10 and 0.5 mM, respectively. The maps were contoured at 5σ. Even when low-pass filtered to 4.2 Å, the density in the selectivity filter vestibule remains a stretch instead of a sphere. Nevertheless, it remains to be investigated whether the stretch of the density observed in the selectivity filter vestibule indeed corresponds to two Ca2+ ions. d, Comparison of Ca2+ coordination by different Cav and Nav channels. The structure of Cav1.1 is superimposed with CavAb26, NavRh28, and TPC169,70 relative to their respective selectivity filters. The PDB accession numbers for CavAb, NavRh, and TPC1 are 4MS2, 4DXW, and 5E1J, respectively. The tentatively assigned Ca2+ ions in Cav1.1 are coloured green and those in the other three indicated channels are coloured wheat.
Extended Data Figure 8 Conformational changes of the intracellular domains.
a, The 3.9 Å map calculated from class Ia particles is almost identical to the 3.6 Å map (class I) except for the better resolution of the β-subunit and the AID motif of α1. b, Distinct conformations of the intracellular domains in class Ia and II reconstructions. The shifts of the β-subunit and the AID motif from class Ia to class II maps are indicated by orange arrows. The adjacent segments in the pore domain also drift to different degrees. For visual clarity, the maps are low-pass filtered to 6 Å. See the Supplementary Video for the morph illustrating the conformational changes of the intracellular elements of the Cav1.1 complex. c, The intracellular III–IV linker of α1 is well resolved. Shown here are class Ia and class II EM maps generated in Chimera. The III–IV linker of α1 forms a short helix that interacts with the carboxyl terminal domain (CTD) of the α1-subunit.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-2 and additional references. (PDF 8382 kb)
Conformational changes of the intracellular segments of the α1 subunit and the β1a subunit
The morph was generated using the two structures built upon Class Ia and Class II maps. Cartoon structures of the α1 (pale blue) and β (pale green) subunits are shown. To generate the morph, the two structures derived from Class Ia and Class II maps were aligned in PyMol. Forty intermediates were then generated using CNS (Brunger, A.T. Version 1.2 of the Crystallography and NMR system. Nat Protoc 2, 2728-33 (2007)). The morphs were merged and displayed in PyMol using "mset" command. (MOV 6568 kb)
Rights and permissions
About this article
Cite this article
Wu, J., Yan, Z., Li, Z. et al. Structure of the voltage-gated calcium channel Cav1.1 at 3.6 Å resolution. Nature 537, 191–196 (2016). https://doi.org/10.1038/nature19321
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19321
This article is cited by
-
Fighting pain: the structure of gabapentin and its binding site in the Cavα2δ subunit
Nature Structural & Molecular Biology (2023)
-
Functional characterization of the transient receptor potential melastatin 2 (TRPM2) cation channel from Nematostella vectensis reconstituted into lipid bilayer
Scientific Reports (2023)
-
EMC chaperone–CaV structure reveals an ion channel assembly intermediate
Nature (2023)
-
Structural basis for CaVα2δ:gabapentin binding
Nature Structural & Molecular Biology (2023)
-
Structure of human NaV1.6 channel reveals Na+ selectivity and pore blockade by 4,9-anhydro-tetrodotoxin
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.