Abstract
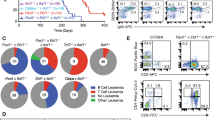
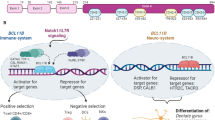
Bcl11a (also called Evi9) functions as a myeloid or B cell proto-oncogene in mice and humans, respectively. Here we show that Bcl11a is essential for postnatal development and normal lymphopoiesis. Bcl11a mutant embryos lack B cells and have alterations in several types of T cells. Phenotypic and expression studies show that Bcl11a functions upstream of the transcription factors Ebf1 and Pax5 in the B cell pathway. Transplantation studies show that these defects in Bcl11a mutant mice are intrinsic to fetal liver precursor cells. Mice transplanted with Bcl11a-deficient cells died from T cell leukemia derived from the host. Thus, Bcl11a may also function as a non-autonomous T cell tumor suppressor gene.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Orkin, S.H. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 1, 57–64 (2000).
Kondo, M., Weissman, I.L. & Akashi, K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91, 661–672 (1997).
Akashi, K., Traver, D., Miyamoto, T. & Weissman, I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197 (2000).
Okuda, T., van Deursen, J., Hiebert, S.W., Grosveld, G. & Downing, J.R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 (1996).
Scott, E.W., Simon, M.C., Anastasi, J. & Singh, H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265, 1573–1577 (1994).
McKercher, S.R. et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15, 5647–5658 (1996).
Georgopoulos, K. et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79, 143–156 (1994).
Bain, G. et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79, 885–892 (1994).
Zhuang, Y., Soriano, P. & Weintraub, H. The helix-loop-helix gene E2A is required for B cell formation. Cell 79, 875–884 (1994).
Lin, H. & Grosschedl, R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376, 263–267 (1995).
Urbanek, P., Wang, Z.Q., Fetka, I., Wagner, E.F. & Busslinger, M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79, 901–912 (1994).
Nutt, S.L., Heavey, B., Rolink, A.G. & Busslinger, M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401, 556–562 (1999).
Rolink, A.G., Nutt, S.L., Melchers, F. & Busslinger, M. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature 401, 603–606 (1999).
Ting, C.N., Olson, M.C., Barton, K.P. & Leiden, J.M. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature 384, 474–478 (1996).
Allen, R.D. 3rd, Bender, T.P. & Siu, G. c-Myb is essential for early T cell development. Genes Dev. 13, 1073–1078 (1999).
Allman, D., Punt, J.A., Izon, D.J., Aster, J.C. & Pear, W.S. An invitation to T and more: notch signaling in lymphopoiesis. Cell 109, S1–S11 (2002).
Li, J. et al. Leukaemia disease genes: large-scale cloning and pathway predictions. Nat. Genet. 23, 348–353 (1999).
Suzuki, T. et al. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 32, 166–174 (2002).
Nakamura, T. et al. Evi9 encodes a novel zinc finger protein that physically interacts with BCL6, a known human B-cell proto-oncogene product. Mol. Cell. Biol. 20, 3178–3186 (2000).
Satterwhite, E. et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood 98, 3413–3420 (2001).
Saiki, Y., Yamazaki, Y., Yoshida, M., Katoh, O. & Nakamura, T. Human EVI9, a homologue of the mouse myeloid leukemia gene, is expressed in the hematopoietic progenitors and down-regulated during myeloid differentiation of HL60 cells. Genomics 70, 387–391 (2000).
Dent, A.L., Shaffer, A.L., Yu, X., Allman, D. & Staudt, L.M. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 276, 589–592 (1997).
Avram, D. et al. Isolation of a novel family of C2H2 zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J. Biol. Chem. 275, 10315–10322 (2000).
Hardy, R.R. & Hayakawa, K. B cell development pathways. Annu. Rev. Immunol. 19, 595–621 (2001).
Li, Y.S., Wasserman, R., Hayakawa, K. & Hardy, R.R. Identification of the earliest B lineage stage in mouse bone marrow. Immunity 5, 527–535 (1996).
Borowski, C. et al. On the brink of becoming a T cell. Curr. Opin. Immunol. 14, 200–206 (2002).
Robey, E. & Fowlkes, B.J. Selective events in T cell development. Annu. Rev. Immunol. 12, 675–705 (1994).
Shortman, K. & Wu, L. Early T lymphocyte progenitors. Annu. Rev. Immunol. 14, 29–47 (1996).
Peault, B., Khazaal, I. & Weissman, I.L. In vitro development of B cells and macrophages from early mouse fetal thymocytes. Eur. J. Immunol. 24, 781–784 (1994).
Kimoto, H. et al. The fetal thymus stores immature hematopoietic cells capable of differentiating into non-T lineage cells constituting the thymus stromal element. Int. Immunol. 5, 1535–1540 (1993).
Ikuta, K. et al. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell 62, 863–8674 (1990).
Kronenberg, M. et al. Rearrangement and transcription of the β-chain genes of the T-cell antigen receptor in different types of murine lymphocytes. Nature 313, 647–653 (1985).
Artavanis-Tsakonas, S., Rand, M.D. & Lake, R.J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999).
Felli, M.P. et al. Expression pattern of Notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand-receptor interactions in intrathymic T cell development. Int. Immunol. 11, 1017–1025 (1999).
Ellisen, L.W. et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66, 649–661 (1991).
Pear, W.S. et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183, 2283–2291 (1996).
Pui, J.C. et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11, 299–308 (1999).
Radtke, F. et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10, 547–558 (1999).
Wilson, A., MacDonald, H.R. & Radtke, F. Notch 1–deficient common lymphoid precursors adopt αβ cell fate in the thymus. J. Exp. Med. 194, 1003–1012 (2001).
Koch, U. et al. Subversion of the T/B lineage decision in the thymus by lunatic fringe-mediated inhibition of Notch-1. Immunity 15, 225–236 (2001).
Izon, D.J. et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 16, 231–243 (2002).
Jehn, B.M., Bielke, W., Pear, W.S. & Osborne, B.A. Cutting edge: protective effects of notch-1 on TCR-induced apoptosis. J. Immunol. 162, 635–638 (1999).
Schebesta, M., Heavey, B. & Busslinger, M. Transcriptional control of B-cell development. Curr. Opin. Immunol. 14, 216–223 (2002).
Kawamata, S., Du, C., Li, K. & Lavau, C. Notch1 perturbation of hemopoiesis involves non-cell-autonomous modifications. J. Immunol. 168, 1738–1745 (2002).
Robey, E. et al. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell 87, 483–492 (1996).
Deftos, M.L., Huang, E., Ojala, E.W., Forbush, K.A. & Bevan, M.J. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 13, 73–84 (2000).
Washburn, T. et al. Notch activity influences the αβ versus γδ T cell lineage decision. Cell 88, 833–843 (1997).
Matzuk, M.M., Finegold, M.J., Su, J.G., Hsueh, A.J. & Bradley, A. α-Inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature 360, 313–319 (1992).
ten Boekel, E., Melchers, F. & Rolink, A. The status of Ig loci rearrangements in single cells from different stages of B cell development. Int. Immunol. 7, 1013–1019 (1995).
Chang, Y., Paige, C.J. & Wu, G.E. Enumeration and characterization of DJH structures in mouse fetal liver. EMBO J. 11, 1891–1899 (1992).
Rolink, A., Haasner, D., Nishikawa, S. & Melchers, F. Changes in frequencies of clonable pre B cells during life in different lymphoid organs of mice. Blood 81, 2290–2300 (1993).
Acknowledgements
We thank S. Spence and T. Kuwata for reviewing the manuscript; D. Swing for help with mouse breeding; J. Wine for tail vein injection; K. Noer and R. Matthai for help with flow cytometry; and the Publication Department of NCI Frederick for graphic illustration. This work was supported by the NCI, Department of Health and Human Services (P.L., N.A.J., N.G.C.). This publication has been funded in whole or in part with federal funds from the NCI, NIH, under contract number NO1-CO-12400 (J.R.K., M.O.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products or organizations imply endorsement by the US Government.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Liu, P., Keller, J., Ortiz, M. et al. Bcl11a is essential for normal lymphoid development. Nat Immunol 4, 525–532 (2003). https://doi.org/10.1038/ni925
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni925
This article is cited by
-
Identification of a genomic DNA sequence that quantitatively modulates KLF1 transcription factor expression in differentiating human hematopoietic cells
Scientific Reports (2023)
-
RNA-seq analysis reveals candidate genes associated with proliferation, invasion, and migration in BCL11A knockdown B-NHL cell lines
Annals of Hematology (2023)
-
APEX1 Nuclease and Redox Functions are Both Essential for Adult Mouse Hematopoietic Stem and Progenitor Cells
Stem Cell Reviews and Reports (2023)
-
CRISPR–Cas9-mediated gene editing of the BCL11A enhancer for pediatric β0/β0 transfusion-dependent β-thalassemia
Nature Medicine (2022)
-
Down-regulation of the transcriptional repressor ZNF802 (JAZF1) reactivates fetal hemoglobin in β0-thalassemia/HbE
Scientific Reports (2022)