Abstract
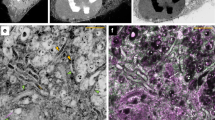
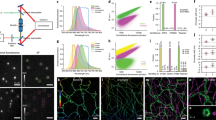
A complete portrait of a cell requires a detailed description of its molecular topography: proteins must be linked to particular organelles. Immunocytochemical electron microscopy can reveal locations of proteins with nanometer resolution but is limited by the quality of fixation, the paucity of antibodies and the inaccessibility of antigens. Here we describe correlative fluorescence electron microscopy for the nanoscopic localization of proteins in electron micrographs. We tagged proteins with the fluorescent proteins Citrine or tdEos and expressed them in Caenorhabditis elegans, fixed the worms and embedded them in plastic. We imaged the tagged proteins from ultrathin sections using stimulated emission depletion (STED) microscopy or photoactivated localization microscopy (PALM). Fluorescence correlated with organelles imaged in electron micrographs from the same sections. We used these methods to localize histones, a mitochondrial protein and a presynaptic dense projection protein in electron micrographs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Cox, G. & Sheppard, C.J. Practical limits of resolution in confocal and non-linear microscopy. Microsc. Res. Tech. 63, 18–22 (2004).
Hell, S.W. Far-field optical nanoscopy. Science 316, 1153–1158 (2007).
Hell, S.W. & Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782 (1994).
Klar, T.A., Jakobs, S., Dyba, M., Egner, A. & Hell, S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. USA 97, 8206–8210 (2000).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Rust, M.J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
Hess, S.T., Girirajan, T.P.K. & Mason, M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006).
Roth, J., Bendayan, M., Carlemalm, E., Villiger, W. & Garavito, M. Enhancement of structural preservation and immunocytochemical staining in low temperature embedded pancreatic tissue. J. Histochem. Cytochem. 29, 663–671 (1981).
Rostaing, P., Weimer, R.M., Jorgensen, E.M., Triller, A. & Bessereau, J. Preservation of immunoreactivity and fine structure of adult C. elegans tissues using high-pressure freezing. J. Histochem. Cytochem. 52, 1–12 (2004).
Morphew, M.K. 3D immunolocalization with plastic sections. Methods Cell Biol. 79, 493–513 (2007).
Murphy, R.M. et al. Size and structure of antigen-antibody complexes. Electron microscopy and light scattering studies. Biophys. J. 54, 45–56 (1988).
Sims, P.A. & Hardin, J.D. Fluorescence-integrated transmission electron microscopy images: integrating fluorescence microscopy with transmission electron microscopy. Methods Mol. Biol. 369, 291–308 (2007).
Micheva, K. & Smith, S. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron 55, 25–36 (2007).
Tsien, R. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998).
Mello, C.C., Kramer, J.M., Stinchcomb, D. & Ambros, V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991).
Frøkjaer-Jensen, C. et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375–1383 (2008).
Yeh, E., Kawano, T., Weimer, R.M., Bessereau, J. & Zhen, M. Identification of genes involved in synaptogenesis using a fluorescent active zone marker in Caenorhabditis elegans. J. Neurosci. 25, 3833–3841 (2005).
Riemersma, J.C. Osmium tetroxide fixation of lipids for electron microscopy. A possible reaction mechanism. Biochim. Biophys. Acta 152, 718–727 (1968).
Clancy, B. & Cauller, L.J. Reduction of background autofluorescence in brain sections following immersion in sodium borohydride. J. Neurosci. Methods 83, 97–102 (1998).
Newman, G.R. & Hobot, J.A. Resin Microscopy and On-Section Immuno-cytochemistry (Springer-Verlag, Berlin, 1993).
Thompson, R. Precise nanometer localization snalysis for individual fluorescent probes. Biophys. J. 82, 2775–2783 (2002).
Yguerabide, J. & Yguerabide, E.E. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications: II. Experimental characterization. Anal. Biochem. 262, 157–176 (1998).
Goldstein, J. et al. Scanning Electron Microscopy and X-Ray Microanalysis (Springer Science and Business Media, LLS, New York, 2003).
Hell, S.W., Lindek, S., Cremer, C. & Stelzer, E.H.K. Measurement of the 4Pi-confocal point spread function proves 75 nm axial resolution. Appl. Phys. Lett. 64, 1335–1337 (1994).
Schmidt, R. et al. Mitochondrial cristae revealed with focused light. Nano Lett. 9, 2508–2510 (2009).
Shtengel, G. et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc. Natl. Acad. Sci. USA 106, 3125–3130 (2009).
Punge, A. et al. 3D reconstruction of high-resolution STED microscope images. Microsc. Res. Tech. 71, 644–650 (2008).
Kanaji, S., Iwahashi, J., Kida, Y., Sakaguchi, M. & Mihara, K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151, 277–288 (2000).
Clark, S.G., Lu, X. & Horvitz, H.R. The Caenorhabditis elegans locus Lin-15, a negative regulator of a tyrosine kinase dignaling pathway, encodes two different proteins. Genetics 137, 987–997 (1994).
Weibull, C. & Christiansson, A. Extraction of proteins and membrane lipids during low temperature embedding of biological material for electron microscopy. J. Microsc. 142, 79–86 (1986).
Acknowledgements
We thank H. Hess and E. Betzig (Janelia Farm) for access to the PALM microscope for proof-of-principle experiments; R. Fetter for sharing protocols, reagents and encouragement; M. Davidson (Florida State University), G. Seydoux (Johns Hopkins Univeristy), S. Eimer (European Neuroscience Institute), R. Leube (Universität Aachen), K. Nehrke (University of Rochester), C. Frøkjær-Jensen (Utah), A. Ada-Nguema (Utah) and M. Hammarlund (Yale University) for DNA constructs. We thank Marine Biological Laboratory for equipment and funding for pilot experiments and Carl Zeiss Inc. for providing access to a beta version of the PAL-M. This research was supported by the US National Institutes of Health (NS034307), National Science Foundation (0920069) and Marine Biology Laboratory fellowship. (The Dart Neuroscience Scholars Program in Learning and Memory).
Author information
Authors and Affiliations
Contributions
S.W. and E.M.J. conceived and designed experiments. G.H., R.J.H. and M.W.D. provided strains and advice. S.W. optimized the methods, prepared the samples and performed PALM imaging. A.P. and K.I.W. performed STED imaging. S.W., S.W.H. and E.M.J. wrote the manuscript. S.W.H. and E.M.J. provided funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Table 1, Supplementary Notes 1–5 (PDF 5940 kb)
Rights and permissions
About this article
Cite this article
Watanabe, S., Punge, A., Hollopeter, G. et al. Protein localization in electron micrographs using fluorescence nanoscopy. Nat Methods 8, 80–84 (2011). https://doi.org/10.1038/nmeth.1537
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1537
This article is cited by
-
Parallel gold enhancement of quantum dots 565/655 for double-labelling correlative light and electron microscopy on human autopsied samples
Scientific Reports (2022)
-
Reactive oxygen FIB spin milling enables correlative workflow for 3D super-resolution light microscopy and serial FIB/SEM of cultured cells
Scientific Reports (2021)
-
Photonic-chip assisted correlative light and electron microscopy
Communications Biology (2020)
-
mEosEM withstands osmium staining and Epon embedding for super-resolution CLEM
Nature Methods (2020)
-
Bioanalysis in single cells: current advances and challenges
Science China Chemistry (2020)