Abstract
Alzheimer’s disease (AD) is a circadian clock-related disease. However, it is not very clear whether pre-symptomatic AD leads to circadian disruption or whether malfunction of circadian rhythms exerts influence on development of AD. Here, we report a functional clock that exists in the hippocampus. This oscillator both receives input signals and maintains the cycling of the hippocampal Per2 gene. One of the potential inputs to the oscillator is orexin signaling, which can shorten the hippocampal clock period and thereby regulate the expression of clock-controlled-genes (CCGs). A 24-h time course qPCR analysis followed by a JTK_CYCLE algorithm analysis indicated that a number of AD-risk genes are potential CCGs in the hippocampus. Specifically, we found that Bace1 and Bace2, which are related to the production of the amyloid-beta peptide, are CCGs. BACE1 is inhibited by E4BP4, a repressor of D-box genes, while BACE2 is activated by CLOCK:BMAL1. Finally, we observed alterations in the rhythmic expression patterns of Bace2 and ApoE in the hippocampus of aged APP/PS1dE9 mice. Our results therefore indicate that there is a circadian oscillator in the hippocampus whose oscillation could be regulated by orexins. Hence, orexin signaling regulates both the hippocampal clock and the circadian oscillation of AD-risk genes.
Similar content being viewed by others
Introduction
Recent reports have revealed that circadian genes are strongly associated with Alzheimer’s disease (AD)1. Researchers have found that circadian rhythms are significantly disturbed in AD and that such disturbance is of significant clinical importance in terms of behavioral symptoms2,3,4,5. Molecular clocks located throughout the body in peripheral tissues and cells are organized into a hierarchical system that is ultimately controlled by a master clock located in the suprachiasmatic nucleus (SCN) of the hypothalamus6,7,8,9,10,11. Autonomous circadian rhythms are generated by intracellular transcriptional feedback loops that feature cis-regulatory elements such as E-boxes, D-boxes, and ROR-elements (ROREs). In general, the so-called clock control genes (CCGs) with these cis-regulatory elements in their promoter regions are regulated by transcriptional activators or repressors7,12,13,14,15.
The deterioration of sleep-wake patterns that results from disturbances in the circadian clock represent some of the most common complaints in elderly human populations, especially in patients with dementia and AD16,17. AD patients are commonly characterized by the aggregation of the pathogenic amyloid-beta (Aβ) peptide and Tau proteins in the brain18,19, especially in the hippocampus and cortex regions of the brain. Accumulating evidence has established that the aberrant expression of core clock genes is strongly associated with the pathogenesis of AD4,15. It is known that the brain-specific knockout of Bmal1 results in AD-like neurodegeneration in mice4,20. Polymorphisms in the CLOCK gene have been associated with the development of AD in humans21,22. Rhythmic expression of BMAL1, CRY1, and PER1 are lost in pineal from both preclinical and clinical AD patients5. Expression of Per2 has also been reported to be a blunted diurnal variation pattern in the SCN in old AD mice23.
Orexin is a neuropeptide hormone encoded by the orexin precursor gene and synthesized in neurons that originate in the lateral hypothalamus (LH). There are two orexin neuropeptides: orexin A and orexin B (OR-A and OR-B). Both of these peptides can bind to two G-protein coupled receptors, orexin receptor 1 and orexin receptor 2, which are encoded, respectively, by Hcrtr1 and Hcrtr224,25. This neuropeptide system plays an important role in numerous behavioral and regulatory functions, including sleep homeostasis and feeding behaviors26,27. Sleep homeostasis is thought to be crucial to hippocampal-dependent memory formation and consolidation28,29. Disruption of sleep homeostasis is known to be directly linked to pathological deterioration in AD. A direct piece of evidence linking orexins and AD is the finding that patients with AD showed altered orexin A levels in cerebrospinal fluid (CSF) relative to normal control individuals2,30. Further, knockout of the orexin precursor gene has been shown to reduce the deposition of Aβ in the in hippocampus and cortex of APP/PS1dE9 mice3. Orexin receptors have also been demonstrated to exert a neuroprotective effect in AD via heterodimerization with GPR103, another G-protein coupled receptor24.
It has been verified that hippocampus-dependent memory impairment is caused mainly by the accumulation of the Aβ peptide and Tau proteins, both in aging and in AD31,32. The physiological isoform of Aβ originates from the amyloid precursor protein (APP) via sequential cleavages that are catalyzed by BACE1 and BACE2 and by Presenilin-1 and Presenilin-2 (PSEN1 and PSEN2). The mechanism of Aβ aggregation has been studied in detail. Recently, the metabolism of Aβ has attracted extensive research attention alongside the re-discovery of the critical function of the APOE gene in AD pathology33,34,35,36. Aβ levels are known to have a diurnal oscillating pattern that has been found to dynamically correlate with the levels of orexins in CSF3,20,37,38. It is also known that the amount of Aβ in CSF increases significantly in the brains of mice during both acute sleep deprivation and following orexin A infusion38. It is typically thought that sleep can accelerate the circulation of CSF, leading to a decrease in Aβ levels37. However, given that the metabolism of Aβ includes not only its clearance, but also its production and transport, we have for some time suspected that the production and transport of Aβ is related both to circadian rhythms and to orexin signaling.
It remains controversial as to whether or not a clock oscillator exists in the hippocampus18,39,40,41,42. However, the reported circadian oscillations of the cAMP/MAPK/CREB signaling pathway strongly suggest that there is indeed an oscillator functioning in the hippocampus43,44,45. Other researchers have also reported rhythmicity in the expression patterns of core clock genes in the hippocampus41,46,47,48. In this study, using real-time recording of hippocampal slices cultured ex vivo combined with pharmacological, genetic, biochemical, and molecular approaches, we confirmed the hypothesis that there is a self-sustained circadian clock in the hippocampus. We also found that the hippocampal clock is a functional clock that can be regulated by inputs such as orexins. Furthermore, we observed that this clock functions to control the transcription of AD-risk genes and that the circadian clock is disturbed by the AD pathology in APP/PS1dE9 mice. Our results suggest that the pathology of AD is associated with the circadian clock in the hippocampus and further suggest that orexin signaling may have an impact on the production and transport of the AD-related Aβ peptide.
Materials and Methods
Animals
All mice used in this paper were housed at 22 ± 2 °C, with 60 ± 5% humidity, and maintained with a LD 12:12 photoperiod (12 h light, 12 h dark, lights on at 07:00). Mice were fed a normal diet and provided water ad libitum. Clockdelta19/+ mice49 and homozygous mPer2::luciferase knock-in mice (mPer2luc/luc)50 were purchased from the Jackson Laboratory. Clockdelta19/+ mice were crossed to mPer2luc/luc reporter mice. From heterozygous offspring, we created double homozygous Clockdelta19/delta19; mPer2luc/luc mice. In this study, APP/PS1dE9 transgenic mice were used to evaluate the mechanism through which the circadian clock contributes to AD51. These mice express a chimeric mouse/human APP (Mo/HuAPP695swe) and a mutant human PSEN1 (PS1-dE9). APP/PS1dE9 mice were also crossed with mPer2luc/luc reporter mice to create APP/PS1dE9; mPer2luc/luc mice. mPer2luc/luc, Clockdelta19/delta19; mPer2luc/luc, and APP/PS1dE9; mPer2luc/luc mice were generated for hippocampal dissection and real-time recoding of the hippocampal oscillation. All experiments for this study were carried out with 2–4 month old male mice, except as otherwise noted. Wild-type (WT) mice were maintained in a LD 12:12 photoperiod condition with free access to food and water for 2 weeks before being kept in complete darkness (DD) for an additional 48 h. WT Mice (n = 3) were sacrificed every 4 h throughout the course of one circadian cycle (both under LD and DD condition). The hippocampus were dissected quickly from brains. Hypothalamus samples were collected every 6 h for one circadian cycle from young (age 4 months, n = 3–5) or aged (aged 12–15 months, n = 3–5) WT and APP/PS1dE9 transgenic mice brains. Animal experiments were performed in accordance with the NIBS institutional regulations, after approval by the Institutional Animal Care and Use Committee (IACUC).
Preparation of hippocampus slices
mPer2luc/luc, Clockdelta19/delta19; mPer2luc/luc, and APP/PS1dE9; mPer2luc/luc mice were anesthetized with 2,2,2-Tribromoethanol (Sigma) and sacrificed at ZT12-15 to reveal the bioluminescence rhythm in the hippocampus; these protocols were performed as previously described49,50. The brain was rapidly removed from the mouse and placed in ice-cold Hanks’ balanced salt solution (HBSS) (Thermo Fisher, pH = 7.2–7.4). The brain was then cut into slices of 220 μm thickness with a vibrating-blade microtome (VT1000S, Leica Microsystems). The slices were maintained in ice-cold HBSS during this procedure until the point when explants were placed into the experimental medium for luciferase recording. The hippocampus was carefully and quickly isolated from the brain slices using scalpels and was then explanted onto a culture membrane (Milli-CM 0.4 μm, EMD Millipore) on top of the liquid surface of a 35 mm Petri dish (Corning) and sealed with a greased 40 mm coverslip. Samples were then cultured with 1.3 mL of HEPES-buffered explant medium supplemented with 1 μM luciferin (Promega) and B-27 supplements (Thermo Fisher). The explants were incubated at 36 °C, and bioluminescence was monitored for one minute in each 10-minute interval using a dish-type luminometer (Actimetrics). The assessment of circadian periods and phases were performed as described in previous reports49,50,51.
Cell culture and transfection
HEK293 cells were grown in regular DMEM supplemented with 10% FBS (Hyclone, GE Healthcare Life Sciences) and antibiotics at 37 °C, 5% CO2. For transfection, rapidly growing cells were trypsinized and re-suspended in DMEM containing 10% FBS lacking antibiotics at a 0.1 × 106 cells/ml concentration. We next added 50 μl of transfection reagent mixture (0.5 μl/well Lipofectamine 2000 in Opti-MEM; Thermo Fisher) to wells containing pre-spotted plasmids. We incubated the wells at room temperature for 20 min and subsequently added 100 μl of cells (0.1 × 105 cells/well). Approximately 6 h after transfection, we replaced this medium with 150 μl of pre-warmed fresh DMEM containing 10% FBS and antibiotics and allowed the cells to grow for an additional 24–30 h. 36 h post-transfection, we replaced this medium with 150 μl of HEPES-buffered explant medium supplemented with luciferin (1 μM) and B-27 supplements; the plates were sealed with an optically clear film. We next loaded these plates into a 36 °C incubator and recorded bioluminescence expression with an Infinite® 200 PRO series microplate reader (Tecan, Thermo Fisher).
Plasmid DNA and materials
The hippocampal slices were treated with final concentrations of 10, 50, and 100 nM orexin A (Abcam) dissolved in DMSO. Forskolin (Sigma) was dissolved in DMSO and the hippocampal slices were treated with a final concentration of 10 μM. Orexin B (Genscript) was dissolved in DMSO; the hippocampal slices were treated with a final concentration of 500 nM. EMPA (Sigma), a high-affinity, reversible, and selective Hcrtr2 antagonist, was dissolved in DMSO52; the hippocampal slices were treated with a final concentration of 10 μM. All compounds, drugs and peptides were titrated in the explant medium to the final concentration and then the prepared medium was added to the 35-mm Petri dish with the slices on the insert. To express E4BP4, the coding sequence of the E4BP4 gene (NM_001289999.1) was amplified from cDNA and subcloned into the pcDNA3.1 plasmid (Thermo Fisher). The human 1.0-kb BACE1-promoter (NC_000011.10) and the 1.4-kb BACE2-promoter (NC_000021.9) were amplified from DNA extracted from HEK293 cells; these amplification products were cloned as pGL3-basic plasmid reporter constructs (Promega) and named, respectively, P(BACE1)-luc and P(BACE2)-luc. The primers used for the PCR amplification of target sequences are detailed in Supplementary Table 4.
RNA isolation and quantitative real-time PCR
Total RNA was extracted from the hippocampus and hypothalamus using Trizol reagent (Thermo Fisher). A 500 ng aliquot of total RNA was reverse transcribed into cDNA using PrimeScript™ RT Master Mix (Takara) and then analyzed with SYBR GREEN qPCR mix (Kapa Biosystems) using a CFX96 instrument (Bio-Rad). The relative quantification of expression levels was performed using a previously-described ΔΔCT calculation method53. Beta-Actin was used as a reference gene. The specific primer pairs used for the analysis of the core clock genes and the AD-risk genes were designed using Primer3 (Supplementary Tables 1–3).
Statistical analysis
OriginPro 2016 software (OriginLab) was used for statistical analyses. Period change data were analyzed with one-way/two-way ANOVA followed by Tukey’s HSD test. To validate whether or not the AD-risk genes displayed circadian oscillations under the LD and DD conditions in the hippocampus, we measured 24 h oscillations in transcript abundance using the JTK_CYCLE algorithm; we set a 5% false discovery rate for detection1,54. We have here reported the results as means with the standard error of the mean (mean ± s.e.m.), and have used P < 0.05 as the criterion for evaluating the inferential statistical significance of differences.
Immunofluorescence
Mice were anesthetized with 2,2,2-Tribromoethanol and fixed by perfusion of 4% paraformaldehyde. Whole brains were dissected and post-fixated with 4% paraformaldehyde for an additional 2 h before dehydration in a 30% sucrose solution overnight at room temperature. Thoroughly dehydrated whole brains were frozen in powdered dry ice. Brain tissues were sliced using a cryostat instrument (CM3050S, Leica) to a thickness of 45 μm. The sections were stained with anti-orexin A (1:125, Santa Cruz) overnight at 4 °C, then incubated with Alexa Fluor 594 labeled anti-goat IgG (1:500, Thermo Fisher) and DAPI (Sigma, 1 mg/mL, 1:1000) for 1 h at room temperature in darkness. Images were obtained via confocal microscopy (Zeiss confocal LSM800). To visualize fibrillary amyloid plaques, sections were stained with Thioflavin T55,56. Images were obtained with a virtual scanning system for microscopy slides (Olympus VS120).
Results
A self-sustained circadian oscillator exists in the hippocampus
We first evaluated whether there was a self-sustained circadian oscillator in the hippocampus. The hippocampus of mPer2luc/luc mice, which we were able to isolate completely from other brain regions (Fig. 1A), clearly maintains oscillations in ex vivo culture (Fig. 1B–D). We found that the oscillations could be synchronized by changing the growth medium and by treatment with forskolin, which activates adenylyl cyclase and then triggers cyclic AMP signaling (Fig. 1C,D). After one week in ex vivo culture, the hippocampal oscillation is dampened and becomes de-synchronized (Fig. 1C,D). This oscillation could be synchronized by treatment with 10 μm forskolin. We also found that Clock malfunction results in defects in maintaining the hippocampal oscillator. Clockdelta19/delta19; mPer2luc/luc mice carry a mutation in the Clock gene that results in a dominant-negative protein that cannot activate transcription57. We dissected the hippocampus and recorded the oscillation of the Clockdelta19/delta19; mPer2luc/luc mice. We found that deficiency in Clock led to an attenuation of circadian rhythms in the hippocampus (Fig. 1B).
A self-sustained circadian oscillator exists in the hippocampus.
(A) Dissected hippocampus slices in ice-cold 1 × HBSS buffer (PH = 7.2–7.4). (B) Deficiency of Clock leads to the attenuation of circadian rhythms in the hippocampus. We recorded the real-time luciferase activity of Clockdelta19/delta19; mPer2luc/luc and WT control hippocampus slices and found an attenuation of circadian rhythms in Clock mutants. (C) The oscillation of the hippocampus slices was damped after one week in ex vivo culture conditions. However, medium changes at days 7–8 induced synchronization of hippocampal oscillation (D) Forskolin (10 μM) treatment induced synchronization of hippocampal oscillation.
To confirm our finding that a self-sustained circadian oscillator exists in the hippocampus, qPCR analysis was conducted to measure the expression levels of core clock genes in the hippocampus in the DD condition. We used the JTK_CYCLE algorithm to characterize cycling variables, including period, phase, and amplitude, for the core clock genes. The expression of 6 out of 7 core clock genes tested here (with the exception being Cry1) exhibited circadian rhythms (Fig. 2B and Table 1), indicating that an intact oscillator exists in the hippocampus. The JTK_CYCLE algorithm results revealed an exact 24 h period for the expression of core clock genes in the hippocampus (Table 1). The mRNA acrophase of Clock and Bmal1, as predicted by the JTK_CYCLE algorithm, occurred at CT0 and CT6, respectively; the JTK_CYCLE algorithm-predicted mRNA acrophase of Per1, Per2, Dbp, and Cry2 occurred, respectively, at CT10, CT14, CT0, and CT10 (Table 1).
qPCR and JTK_CYCLE algorithm analysis of core clock gene in hippocampus.
The qPCR results and the cycling parameters predicted by the JTK_CYCLE algorithm of core clock genes under the DD condition in the hippocampus. (A) Schedule for sampling in the LD and DD conditions. WT mice were decapitated at ZT4, 8, 12, 16, 20, and 24 under the LD condition or at CT0, 4, 8, 12, 16, and 20 under the DD condition; (B) Gene expression patterns of core clock genes in the hippocampus in the DD condition (Dbp, Bmal1, Per1, Per2, Clock, and Cry2). The qPCR primers used here are listed in Supplementary Tables 2 and 3.
Treatment with orexins shortens the period of the hippocampal oscillator
Intact circadian oscillators can be regulated by input signals such as hormones, metabolites, and neuropeptides7. We evaluated whether oscillation in the hippocampus can be regulated by such signals. Orexins are essential regulators of sleep/wake cycles and are typically thought to be involved in the circadian clock, hippocampus-dependent social memory, and Aβ pathology. We therefore hypothesized that orexin signaling might be able to regulate the hippocampal oscillator. As expected, we found that treatment with orexin A or B (OR-A and OR-B, for short) did indeed regulate the hippocampal clock, leading to a shortened period of hippocampal oscillation in ex vivo culture (Fig. 3A). The period of explants was 23.68 ± 0.12 h with 100 nM OR-A treatment and was 24.4 ± 0.09 h with control DMSO treatment (Fig. 3B, Supplemental Fig. 1A–C). Treatment with 500 nM OR-B also shortened the period of hippocampal oscillation ex vivo (Fig. 3C). The period of explants was 23.57 ± 0.08 h with 500 nM orexin B treatment and was 24.54 ± 0.03 h with control DMSO treatment. Treatment with 10 μM forskolin did not alter the period of the hippocampal oscillator (Fig. 3C,D).
Orexin treatment led to shortening of the period of the hippocampal oscillator.
(A) Effects of orexin A and DMSO treatments on hippocampal oscillation. (B) Orexin A treatment induced shortening of the period of the hippocampal oscillator as compared with the no-treatment control, the DMSO control, and the Forskolin (10 μM) treatment (n = 4–9); Bar graph (mean ± s.e.m.) of period; length of periods was analyzed by two-factor ANOVA, followed by Tukey’s HSD *P < 0.05. (C) Effects of orexin B and DMSO treatments on hippocampal oscillation. (D) Orexin B treatment induced shortening of the period of the hippocampal oscillator as compared with the DMSO control and the Forskolin (10 μM) treatment (n = 4–18); Bar graph (mean ± s.e.m.) of period; length of periods was analyzed by one-way ANOVA, followed by Tukey’s HSD *P < 0.05. (E,F,H,G) EMPA blocks the orexin A-induced shortening of the period of the hippocampal oscillator. The period showed no difference before EPMA and orexin A treatment. (E) EMPA blocks the orexin A-induced shortening of the period of the hippocampal oscillator. (F,G) Expression analysis of the orexin receptors shows that Hcrtr2 is the major receptor in the hippocampus.
qPCR analyses of orexin receptor genes revealed that Hcrtr2 levels are higher than Hcrtr1 levels in the hippocampus, suggesting that Hcrtr2 is likely the primary receptor for orexins in this brain region (Fig. 3G). To block the binding of orexins to the receptors, we performed co-treatment experiments with hippocampus slices that tested combinations of EMPA and OR-A or OR-B. In the presence of EMPA, both OR-A and OR-B completely failed to shorten the period of the hippocampal clock (Fig. 3E,F,H; Supplemental Fig. 1D,E). All of these results demonstrate that both the OR-A peptide and the OR-B peptide can function, in redundant roles, in shortening the hippocampal clock.
Circadian oscillation of Alzheimer’s disease-risk genes
The key AD-risk gene Psen1 has previously been shown to have rhythmic expression in the liver, which is thought to possess a functional circadian oscillator1. Since AD occurs in the hippocampus and the cerebral cortex of the brain, not in peripheral organs, we sought to measure whether, or how, the clock regulates the expression of AD-risk genes in the hippocampus. We therefore monitored the expression of a group of 77 candidate genes (AD RT2 Profiler PCR Array, QIAGEN)58,59 that are known to be involved in most of the cellular AD-related signaling pathways (Supplementary Table 1) to determine whether these genes merit classification as CCGs. The JTK_CYCLE algorithm was used to identify and characterize the cycling variables of the qPCR dataset for these 77 candidate genes. We found that nearly half of the AD-risk genes exhibited rhythmic expression patterns. We found that 39 and 34 of the candidate genes had rhythmic expression patterns in the LD condition and the DD condition, respectively (Fig. 4A). To narrow down the size of our target gene list, we concatenated the genes that were classified by JTK_CYCLE to be CCGs under both LD and DD conditions. We found that at least 12 out of 77 AD-risk genes are under circadian control in the hippocampus (Fig. 4A; Table 2). Gnb5, Sncβ, Casp3, ApoE, Cdc2, Bace1, Gng1, Psen2, Gsk3α, Apba1, Bace2, and Prkcδ are the genes most likely to be under clock regulation, as the expression of all of these genes was found to be rhythmic under both the LD and DD conditions in the hippocampus, and the statistical confidence levels for all of their predicted JTK_CYCLE algorithm cycling variables were high (Fig. 4B; Table 2).
qPCR results and Venn diagram of commonly rhythmic expressed genes in hippocampus under different light conditions.
(A) Genes that are predicted by the JTK_CYCLE algorithm to be potentially rhythmically expressed under both the LD condition and the DD condition in the hippocampus. (B) The expression patterns of Bace1, Bace2, Psen1, Prkcδ, Gsk3α, and Apba1 in the hippocampus in the LD condition; the cycling parameters predicted by the JTK_CYCLE algorithm are listed under the graph of the relative expression of each gene. The qPCR primers used for the expression analysis of the target genes are detailed in Supplementary Tables 1 and 3.
Bace1 and Bace2 are directly regulated by the hippocampal clock
The expression of both Bace1 and Bace2 was classified as rhythmic in the hippocampus based on our qPCR analysis. We found that the promoter regions of BACE1 and BACE2 contain putative cis-elements that are potential binding sites for core clock genes. There is a putative D-box in the promoter region of BACE1 (Fig. 5A). There are four potential E-boxes in the promoter region of BACE2 (Fig. 5A). We sought to confirm, in vitro, whether the BACE1 and BACE2 promoters could be regulated by core clock transcription activators such as CLOCK:BMAL1 and ROREs or be regulated by a repressor like E4BP4. Our co-transfection experiments showed that P(BACE2)-Luc expression was activated by the CLOCK:BMAL1, and we found that the activation of BACE2 depended strictly on the amount of CLOCK:BMAL1 present (Fig. 5B, right panel). P(BACE1)-Luc expression was inhibited by the E4BP4 repressor (Fig. 5B, left panel). Further qPCR analysis found that Bace2 decreased the expression at ZT5 and ZT17 in Clock mutant mice, as did the canonical E-box gene (Fig. 5C). All these data confirm that Bace1 and Bace2 are potential CCGs and could be directly regulated by the hippocampal clock.
Expression of the BACE genes is controlled by the clock.
(A) Schematic plot of the promoters of the BACE genes, including four possible E-boxes located in the 1.4-kb BACE2 promoter and one D-box located in the 1-kb BACE1 promoter. (B) Left pane: transfection of E4BP4 inhibited the transcription of Bace1 in a dose-dependent manner. Right pane: The CLOCK:BMAL1 complex activated the expression of Bace2 in a dose-dependent manner. (C) The expression of Per2 and Bace2 between Clock mutant and WT mice in hippocampus. Clockdelta19/delta19 and WT mice (n = 3) were sacrificed at ZT5 and ZT7 under the LD condition; the expressions of Per2 and Bace2 were decreased at ZT5 and ZT17 in the Clockdelta19/delta19 mice due to the deficiency of the Clock gene; Bar graph (mean ± s.e.m.) of relative gene expression (n = 3, N.S. P > 0.05; *P < 0.05; **P < 0.01, student’s t-test).
The regulation of E-box genes is crucially important in Alzheimer’s disease
The deposition of the Aβ peptide is a major pathological aspect of AD. We confirmed this phenotype in our AD-model mice (APP/PS1dE9). Thioflavin T (TFT) is a benzothiazole dye that exhibits enhanced fluorescence upon binding to amyloid fibrils60. Our TFT staining results indicated that there were massive amyloid fibrils in the cortex and hippocampus regions of APP/PS1dE9 mice compared with WT controls (Fig. 6A). We next used qPCR analysis to examine the expression patterns of core clock genes and AD-risk genes, and evaluated how AD progression affected the expression profiles of these genes. As predicted, aged AD mice experience changes in the expression of risk genes, including Bace2 and ApoE. Interestingly, a subset of the tested core clock genes of aged APP/PS1dE9 mice, including Per1, Per2, Cry1, and Dbp, had altered expression patterns compared with WT controls. Dbp expression was elevated at ZT5 and ZT17 in the hippocampus of aged APP/PS1dE9 mice, but was decreased at ZT23 compared with WT control; however, the expression of Per1, Per2, and Cry1 was decreased at ZT11 but elevated at ZT23 (Fig. 6D). Furthermore, the expression of Bace2 and ApoE, both of which have been verified by us and by other researchers to be E-box genes that contain non-canonical E-box motifs in their promoter regions61, was also altered in the hippocampus of aged APP/PS1dE9 mice at ZT5, ZT11, and ZT17 (Fig. 6C). Non-E-box-containing core clock genes, including Bmal1 and Clock, showed only minor, if any, changes in their mRNA levels (Supplemental Fig. 3A,C). There were no differences between aged APP/PS1dE9 mice compared with WT controls in the expression of Nr1d1 and Nr1d2 (Supplemental Fig. 3B), two additional E-box genes, possibly because these genes, which are representative of a secondary loop in the circadian network62, are not susceptible to the AD pathology.
The rhythmicity of Bace2 and ApoE expression is altered in the hippocampus of APP/PS1dE9 mice.
(A) Thioflavin T staining of WT and APP/PS1dE9 mice. Fibrillary amyloid plaques existed only in the brains of the APP/PS1dE9 mice (Red asterisks indicate fibrillary amyloid plaques). (B) The diurnal expression pattern of the orexin precursor gene is altered in the hypothalamus of APP/PS1dE9 mice. Expression of orexin precursor mRNA was analyzed by qPCR in the hypothalamus of WT and APP/PS1dE9 mice. The expression of this gene was higher at ZT5 in APP/PS1dE9 mice compared with aging WT mice; we did not detect any differences in mRNA levels at other time points. (C) The rhythmicity of Bace2 and ApoE expression is altered in the hippocampus of APP/PS1dE9 mice. Left panel: expression of Bace2 shows that this AD-related gene increased dramatically in the hippocampus of APP/PS1dE9 mice. Right panel: ApoE gene expression elevated the expression at ZT5, ZT11, and ZT17. The expression of this gene, which is highly correlated with the metabolism of Aβ, increased slightly in the hippocampus of APP/PS1dE9 mice. (D) Canonical E-box genes such as Dbp, Per1, Per2, and Cry1 changed the rhythmicity in the hippocampus of APP/PS1dE9 mice (n = 3–5, N.S. P > 0.05; *P < 0.05; **P < 0.01, student’s t-test). Top left panel: Dbp; Top right panel: Per1; bottom left panel: Per2; bottom right panel: Cry1.
Elevated orexin levels in the hypothalamus of Alzheimer’s disease mice
Finally, we wondered whether the expression of orexins in the brain is governed by the clock. Intriguingly, the expression of the orexin precursor gene was found to be rhythmic in the hypothalamus (Fig. 6B and Supplemental Fig. 2A). Further immunofluorescence experiments confirmed that orexin A exhibits the same diurnal expression pattern at the protein level: In the lateral hypothalamus area, the immunostaining signal for the orexin A peptide is higher at ZT17 (Supplemental Fig. 2B), a finding consistent with our qPCR analysis of the orexin precursor gene (Supplemental Fig. 2A). Interestingly, the expression pattern of the orexin precursor gene is altered in the hypothalamus in APP/PS1dE9 mice. The transcription of this gene is higher at ZT5 in APP/PS1dE9 mice than in WT control mice (Fig. 6B). At other time points, the expression of this gene did not differ significantly between the two mouse genotypes (Fig. 6B).
Discussion
It is well established that the central clock oscillator is located in the SCN. This clock oscillator can run independently and robustly in ex vivo conditions for months50. However, the hypothesis of a single central clock has been challenged by the discovery of self-sustained oscillators in several tissues, including the liver, lung, and kidney. Even in the mammalian brain, it has been reported that circadian oscillation exists in the amygdala, arcuate nucleus, bed nucleus of the stria terminalis, dorsomedial hypothalamus, habenula, lateral hypothalamus, olfactory bulb, pineal gland, and pituitary gland, as well as SCN14. It seems plausible that these peripheral oscillators may be particularly relevant for localized rhythmic events. Here, we confirmed that there is an intact and functional oscillator in the hippocampus. We also observed that Clock deficiency impairs local oscillation in hippocampus. Our real-time recording results for the hippocampal oscillation of Clockdelta19/delta19; mPer2luc/luc mice were in line with reports claiming that the genetic disruption of the Clock impaired local oscillation in the SCN10,57.
An intact and functional oscillator is typically held to include three major components: a transcription and translation feedback loop (TTFL) oscillator, input pathways, and output pathways. Considering that AD patients are known to have sleep/wake abnormalities related to malfunctions in their orexin systems2,3,30,38,63, we tested the effect of orexins on the hippocampal oscillator and evaluated whether orexin signals function as inputs for the clock. We found that orexins are indeed involved in regulating the hippocampal oscillator. Surprisingly, these inputs can speed the hippocampal oscillator, resulting in a shortened period. Consistently, real-time recording of the hippocampal oscillator in the aged APP/PS1dE9; mPer2luc/luc mice revealed that there is indeed a shortened period in these mice (Supplemental Fig. 1F,G). qPCR analysis of the orexin precursor gene also indicated that there were higher orexin levels in the hypothalamus area of aged APP/PS1dE9 mice than in WT control mice (Fig. 6B). All of these results show that a shorter period is a potential intrinsic alteration of the hippocampal circadian oscillator that accompanies the AD pathology.
Phosphorylation of the PERs is known to contribute in the determination of the period length of the circadian clock64,65. Many pathways regulate circadian timing by altering the phosphorylation status of the PERs. For example, phosphorylation at Ser47 in Drosophila PER and dephosphorylation at Ser662 in human PER2 have been verified as period-shortening molecular events66,67. Orexins can bind to selective G-protein-coupled receptors and activate kinases and phosphatases24,27, which may then be involved in regulating the phosphorylation and dephosphorylation of critical amino acid residues in Per2 that result in a shortening of the period of the hippocampal oscillator64,65. Previous studies have shown that a deficiency in the levels of orexins in APP/PS1dE9 mice led to a reduction in the amount of fibrillary amyloid plaque in the cortex and hippocampus as compared to control mice3. We also confirmed that there were modest changes, such as period shortening, of the hippocampal clock in aged APP/PS1dE9 mice (Supplemental Fig. 1F,G), which might have been caused by chronic elevated orexin levels in these mice. Our observations that a higher expression level of the orexin precursor gene accompanies the deterioration of amyloid deposition in APP/PS1dE9 mice, and that high-orexin treatment can shorten the oscillation period of the hippocampal circadian clock, are consistent with the results of these previous studies.
Negative transcription feedback loops are a core mechanism underlying the circadian clock. We questioned whether the transcription of AD-risk genes could be understood as outputs of the hippocampal oscillator. Using the JTK_CYCLE algorithm, a number of AD-risk genes were identified and found to have rhythmic expression profiles. There are two major pathways known to be involved in AD pathology: One includes the genes related to Aβ metabolism (its generation, oligomerization, clearance, and degradation); the other is the pathway for the hyperphosphorylation of the Tau protein33,68. In our study, genes from both of these pathways were observed to have rhythmic expression patterns. Such genes related to Aβ generation, oligomerization, clearance, and degradation include Bace1, Bace2, and ApoE. Such genes related to hyperphosphorylation of the Tau protein include Gsk3α and Prkcδ.
It is known that BACE1 and BACE2 are required for the production of the Aβ peptide; these genes are considered to be central to the pathogenesis of AD. APOE was previously thought to be a cholesterol transporter69,70. Intriguingly, ApoE, which was recently verified as a regulator of Aβ metabolism33,34,71, has rhythmic expression. The rhythmicity of Bace1, Bace2, and ApoE expression has been proposed to contribute to the diurnal pattern of Aβ to at least some extent38,72,73,74. These suppositions are supported by reports that the acrophase of Bace2 coincides with the phase of Aβ in CSF38,75. Although it remains controversial as to whether decreased or increased orexin levels lead to deterioration in AD2,3,30,38, many recent studies have assumed the increasingly common view that, at a minimum, orexin levels are disturbed in AD. Our findings indicate that orexin expression exhibits a diurnal pattern, and our data showing elevated expression levels of the orexin precursor gene at ZT5 in the hypothalamus area of aged APP/PS1dE9 mice have led us to prefer the view that higher orexins levels lead to deterioration in AD. Our observation of higher orexin levels in the hypothalamus area is consistent with previous results showing that A) knockout of the orexin precursor gene in APP/PS1dE9 mice led to decreased amounts of amyloid fibrils3, and B) that higher orexin levels are correlated with higher Aβ levels37,76.
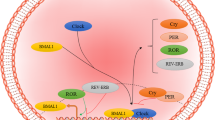
Further expression analysis of the BACEs showed that BACE2 is indeed activated by the CLOCK:BMAL1 complex. The expression of BACE1, which seems to have a substantial phase delay relative to Bace2 expression, is inhibited by E4BP4, another key clock regulator. Additional expression profile data for AD-risk genes in APP/PS1dE9 mice strengthen our view that AD-risk genes are under the control of the hippocampal circadian clock. We analyzed the expression of Bace1, Bace2, ApoE, and Gsk3α in the hippocampus and observed that, compared to WT controls, the expression of Bace2 and ApoE, both of which are E-box genes, was elevated in APP/PS1dE9 mice at ZT5, ZT11, and ZT17. Interestingly, the canonical E-box genes (Dbp, Per1, Per2, and Cry1) also had altered expression patterns in the hippocampus of APP/PS1dE9 mice. Old APP/PS1dE9 mice displayed blunted diurnal variation expression of Per1 and Per2 in the hippocampus. These data were consistent with a previous report on the SCN in AD animals23. Our results imply that E-box genes are susceptible regulatory mechanisms that relate to the pathology of AD. Therefore, we propose the following network: Orexin signaling influences hippocampal oscillation; simultaneously, the hippocampal oscillator controls the circadian expression of AD-risk genes like Bace1, Bace2, and ApoE, which are key genes in Aβ metabolism; the rhythmicity of these AD-risk genes indicates that AD is associated with circadian oscillation; finally, orexin signaling is involved in the reciprocal control of the core clock feedback loop and AD (Fig. 7).
A hypothetical model for the reciprocal control of the hippocampal oscillator.
Orexin signaling and AD-risk genes. (1) and (3) A self-sustained circadian oscillator exists in the hippocampus; (3) and (S1) Orexin signaling can speed up hippocampal oscillation, acting as an input signal to the hippocampal clock; (2) The hippocampal oscillator controls the expression of core clock genes; (4) Several key AD-risk genes, (5) including Bace1/2, which are regulated through D-boxes and E-boxes, respectively; (6) The rhythmicity of Bace2 and ApoE are altered in the hippocampus of APP/PS1dE9 mice; (6) and (S2) The orexin precursor gene is rhythmic in the brain. Thus, our model supports the following notions: Orexin signaling influences hippocampal oscillation; the hippocampal oscillator controls the rhythmic expression of AD-risk genes like Bace1, Bace2, and ApoE, which are the key genes in the metabolism of Aβ; the rhythmicity of these AD-risk genes indicate that disrupted circadian oscillation and sleep both contribute to the risk of AD; and, finally, orexin signaling is involved in reciprocal control of the core clock feedback loop and AD. The blue arrows show the pathways or physiological processes involved in the present study; the black arrows show the well-known pathways or physiological processes; the dashed black arrows represent uncertain relationships between the pathways or physiological processes.
In this study, we confirmed that there is an intact hippocampal oscillator; in other words, the hippocampal oscillator contains the molecular components required for SCN-independent, persistent circadian oscillation. We also demonstrated that orexins, which shorten the period of the hippocampal oscillator, are potential inputs to the oscillator. The transcription of AD-risk genes, including Bace1, Bace2, and ApoE, appear to be outputs of the oscillator. E-box genes such as Bace2, ApoE, Dbp, Per1, and Per2 were found to be more susceptible than genes of other clock subclasses to alterations relating to AD in APP/PS1dE9 mice. In conclusion, orexin signaling regulates the period of the hippocampal oscillator and circadian oscillation of Alzheimer’s disease-risk genes.
Additional Information
How to cite this article: Ma, Z. et al. Orexin signaling regulates both the hippocampal clock and the circadian oscillation of Alzheimer’s disease-risk genes. Sci. Rep. 6, 36035; doi: 10.1038/srep36035 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Zhang, R., Lahens, N. F., Ballance, H. I., Hughes, M. E. & Hogenesch, J. B. A circadian gene expression atlas in mammals: implications for biology and medicine. Proceedings of the National Academy of Sciences of the United States of America 111, 16219–16224 (2014).
Scammell, T. E., Matheson, J. K., Honda, M., Thannickal, T. C. & Siegel, J. M. Coexistence of narcolepsy and Alzheimer’s disease. Neurobiology of aging 33, 1318–1319 (2012).
Roh, J. H. et al. Potential role of orexin and sleep modulation in the pathogenesis of Alzheimer’s disease. The Journal of experimental medicine 211, 2487–2496 (2014).
Musiek, E. S. et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. The Journal of clinical investigation 123, 5389–5400 (2013).
Coogan, A. N. et al. The circadian system in Alzheimer’s disease: disturbances, mechanisms, and opportunities. Biological psychiatry 74, 333–339 (2013).
Doherty, C. J. & Kay, S. A. Circadian control of global gene expression patterns. Annual review of genetics 44, 419–444 (2010).
Welsh, D. K., Takahashi, J. S. & Kay, S. A. Suprachiasmatic nucleus: cell autonomy and network properties. Annual review of physiology 72, 551–577 (2010).
Hughes, A. T. & Piggins, H. D. Disruption of daily rhythms in gene expression: the importance of being synchronised. BioEssays: news and reviews in molecular, cellular and developmental biology 36, 644–648 (2014).
Buhr, E. D. & Takahashi, J. S. Molecular components of the Mammalian circadian clock. Handbook of experimental pharmacology, 3–27 (2013).
Ko, C. H. & Takahashi, J. S. Molecular components of the mammalian circadian clock. Human molecular genetics 15 Spec No 2, R271–R277 (2006).
Huang, W., Ramsey, K. M., Marcheva, B. & Bass, J. Circadian rhythms, sleep, and metabolism. The Journal of clinical investigation 121, 2133–2141 (2011).
Bachman, D. & Rabins, P. “Sundowning” and other temporally associated agitation states in dementia patients. Annual review of medicine 57, 499–511 (2006).
Partch, C. L., Green, C. B. & Takahashi, J. S. Molecular architecture of the mammalian circadian clock. Trends in cell biology 24, 90–99 (2014).
Dibner, C., Schibler, U. & Albrecht, U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual review of physiology 72, 517–549 (2010).
Mattis, J. & Sehgal, A. Circadian Rhythms, Sleep, and Disorders of Aging. Trends in endocrinology and metabolism: TEM (2016).
Ju, Y. E., Lucey, B. P. & Holtzman, D. M. Sleep and Alzheimer disease pathology–a bidirectional relationship. Nature reviews Neurology 10, 115–119 (2014).
Slats, D., Claassen, J. A., Verbeek, M. M. & Overeem, S. Reciprocal interactions between sleep, circadian rhythms and Alzheimer’s disease: focus on the role of hypocretin and melatonin. Ageing research reviews 12, 188–200 (2013).
Hastings, M. H. & Goedert, M. Circadian clocks and neurodegenerative diseases: time to aggregate? Current opinion in neurobiology 23, 880–887 (2013).
Jagust, W. J. & Mormino, E. C. Lifespan brain activity, beta-amyloid, and Alzheimer’s disease. Trends in cognitive sciences 15, 520–526 (2011).
Musiek, E. S., Xiong, D. D. & Holtzman, D. M. Sleep, circadian rhythms, and the pathogenesis of Alzheimer disease. Experimental & molecular medicine 47, e148 (2015).
Chen, Q., Huang, C. Q., Hu, X. Y., Li, S. B. & Zhang, X. M. Functional CLOCK gene rs1554483 G/C polymorphism is associated with susceptibility to Alzheimer’s disease in the Chinese population. The Journal of international medical research 41, 340–346 (2013).
Chen, H. F., Huang, C. Q., You, C., Wang, Z. R. & Si-qing, H. Polymorphism of CLOCK gene rs 4580704 C > G is associated with susceptibility of Alzheimer’s disease in a Chinese population. Archives of medical research 44, 203–207 (2013).
Duncan, M. J. et al. Effects of aging and genotype on circadian rhythms, sleep, and clock gene expression in APPxPS1 knock-in mice, a model for Alzheimer’s disease. Experimental neurology 236, 249–258 (2012).
Davies, J. et al. Orexin receptors exert a neuroprotective effect in Alzheimer’s disease (AD) via heterodimerization with GPR103. Scientific reports 5, 12584 (2015).
Watanabe, Y., Muller, M. K., von Engelhardt, J., Sprengel, R., Seeburg, P. H. & Monyer, H. Age-Dependent Degeneration of Mature Dentate Gyrus Granule Cells Following NMDA Receptor Ablation. Frontiers in molecular neuroscience 8, 87 (2015).
Thompson, M. D., Xhaard, H., Sakurai, T., Rainero, I. & Kukkonen, J. P. OX1 and OX2 orexin/hypocretin receptor pharmacogenetics. Frontiers in neuroscience 8, 57 (2014).
Tsujino, N. & Sakurai, T. Orexin/hypocretin: a neuropeptide at the interface of sleep, energy homeostasis, and reward system. Pharmacological reviews 61, 162–176 (2009).
Yang, G., Lai, C. S., Cichon, J., Ma, L., Li, W. & Gan, W. B. Sleep promotes branch-specific formation of dendritic spines after learning. Science 344, 1173–1178 (2014).
Siegel, J. M. Clues to the functions of mammalian sleep. Nature 437, 1264–1271 (2005).
Fronczek, R. et al. Hypocretin (orexin) loss in Alzheimer’s disease. Neurobiology of aging 33, 1642–1650 (2012).
Mander, B. A. et al. beta-amyloid disrupts human NREM slow waves and related hippocampus-dependent memory consolidation. Nature neuroscience 18, 1051–1057 (2015).
Craig, L. A. & McDonald, R. J. Chronic disruption of circadian rhythms impairs hippocampal memory in the rat. Brain research bulletin 76, 141–151 (2008).
Raichlen, D. A. & Alexander, G. E. Exercise, APOE genotype, and the evolution of the human lifespan. Trends in neurosciences 37, 247–255 (2014).
Kanekiyo, T., Xu, H. & Bu, G. ApoE and Abeta in Alzheimer’s disease: accidental encounters or partners? Neuron 81, 740–754 (2014).
Jack, C. R. Jr. et al. Suspected non-Alzheimer disease pathophysiology-concept and controversy. Nature reviews Neurology 12, 117–124 (2016).
Castellano, J. M. et al. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Science translational medicine 3, 89ra57 (2011).
Xie, L. et al. Sleep drives metabolite clearance from the adult brain. Science 342, 373–377 (2013).
Kang, J. E. et al. Amyloid-beta dynamics are regulated by orexin and the sleep-wake cycle. Science 326, 1005–1007 (2009).
Wang, L. M. et al. Expression of the circadian clock gene Period2 in the hippocampus: possible implications for synaptic plasticity and learned behaviour. ASN neuro 1 (2009).
Chun, L. E., Woodruff, E. R., Morton, S., Hinds, L. R. & Spencer, R. L. Variations in Phase and Amplitude of Rhythmic Clock Gene Expression across Prefrontal Cortex, Hippocampus, Amygdala, and Hypothalamic Paraventricular and Suprachiasmatic Nuclei of Male and Female Rats. Journal of biological rhythms 30, 417–436 (2015).
Wakamatsu, H., Yoshinobu, Y., Aida, R., Moriya, T., Akiyama, M. & Shibata, S. Restricted-feeding-induced anticipatory activity rhythm is associated with a phase-shift of the expression of mPer1 and mPer2 mRNA in the cerebral cortex and hippocampus but not in the suprachiasmatic nucleus of mice. The European journal of neuroscience 13, 1190–1196 (2001).
Abe, M. et al. Circadian rhythms in isolated brain regions. The Journal of neuroscience: the official journal of the Society for Neuroscience 22, 350–356 (2002).
Phan, T. X., Chan, G. C., Sindreu, C. B., Eckel-Mahan, K. L. & Storm, D. R. The diurnal oscillation of MAP (mitogen-activated protein) kinase and adenylyl cyclase activities in the hippocampus depends on the suprachiasmatic nucleus. The Journal of neuroscience: the official journal of the Society for Neuroscience 31, 10640–10647 (2011).
Eckel-Mahan, K. L. et al. Circadian oscillation of hippocampal MAPK activity and cAmp: implications for memory persistence. Nature neuroscience 11, 1074–1082 (2008).
Eckel-Mahan, K. L. Circadian Oscillations within the Hippocampus Support Memory Formation and Persistence. Frontiers in molecular neuroscience 5, 46 (2012).
Shieh, K. R. Distribution of the rhythm-related genes rPERIOD1, rPERIOD2, and rCLOCK, in the rat brain. Neuroscience 118, 831–843 (2003).
Uz, T. et al. Effect of fluoxetine and cocaine on the expression of clock genes in the mouse hippocampus and striatum. Neuroscience 134, 1309–1316 (2005).
Jilg, A. et al. Temporal dynamics of mouse hippocampal clock gene expression support memory processing. Hippocampus 20, 377–388 (2010).
Vitaterna, M. H. et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264, 719–725 (1994).
Yoo, S. H. et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proceedings of the National Academy of Sciences of the United States of America 101, 5339–5346 (2004).
Jankowsky, J. L. et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Human molecular genetics 13, 159–170 (2004).
Ko, C. H. et al. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS biology 8, e1000513 (2010).
Savelyev, S. A., Larsson, K. C., Johansson, A. S. & Lundkvist, G. B. Slice preparation, organotypic tissue culturing and luciferase recording of clock gene activity in the suprachiasmatic nucleus. Journal of visualized experiments: JoVE (2011).
Landgraf, D., Wang, L. L., Diemer, T. & Welsh, D. K. NPAS2 Compensates for Loss of CLOCK in Peripheral Circadian Oscillators. PLoS genetics 12, e1005882 (2016).
Malherbe, P. et al. Biochemical and behavioural characterization of EMPA, a novel high-affinity, selective antagonist for the OX(2) receptor. British journal of pharmacology 156, 1326–1341 (2009).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research 29, e45 (2001).
Hughes, M. E., Hogenesch, J. B. & Kornacker, K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. Journal of biological rhythms 25, 372–380 (2010).
Biancalana, M. & Koide, S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochimica et biophysica acta 1804, 1405–1412 (2010).
Rajamohamedsait, H. B. & Sigurdsson, E. M. Histological staining of amyloid and pre-amyloid peptides and proteins in mouse tissue. Methods Mol Biol 849, 411–424 (2012).
Turek, F. W. et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308, 1043–1045 (2005).
Poitevin, S. et al. Sphingosine kinase 1 expressed by endothelial colony-forming cells has a critical role in their revascularization activity. Cardiovascular research 103, 121–130 (2014).
Chahrour, M. et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229 (2008).
Khurana, R. et al. Mechanism of thioflavin T binding to amyloid fibrils. Journal of structural biology 151, 229–238 (2005).
Salero, E., Gimenez, C. & Zafra, F. Identification of a non-canonical E-box motif as a regulatory element in the proximal promoter region of the apolipoprotein E gene. The Biochemical journal 370, 979–986 (2003).
Zhang, E. E. & Kay, S. A. Clocks not winding down: unravelling circadian networks. Nature reviews Molecular cell biology 11, 764–776 (2010).
Shan, L., Dauvilliers, Y. & Siegel, J. M. Interactions of the histamine and hypocretin systems in CNS disorders. Nature reviews Neurology 11, 401–413 (2015).
Vanselow, K. et al. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes & development 20, 2660–2672 (2006).
Edery, I., Zwiebel, L. J., Dembinska, M. E. & Rosbash, M. Temporal phosphorylation of the Drosophila period protein. Proceedings of the National Academy of Sciences of the United States of America 91, 2260–2264 (1994).
Williams, J. A. & Sehgal, A. Molecular components of the circadian system in Drosophila. Annual review of physiology 63, 729–755 (2001).
Golombek, D. A. & Rosenstein, R. E. Physiology of circadian entrainment. Physiological reviews 90, 1063–1102 (2010).
Martin, L. et al. Tau protein kinases: involvement in Alzheimer’s disease. Ageing research reviews 12, 289–309 (2013).
Vedhachalam, C. et al. The C-terminal lipid-binding domain of apolipoprotein E is a highly efficient mediator of ABCA1-dependent cholesterol efflux that promotes the assembly of high-density lipoproteins. Biochemistry 46, 2583–2593 (2007).
Yokoyama, S., Kawai, Y., Tajima, S. & Yamamoto, A. Behavior of human apolipoprotein E in aqueous solutions and at interfaces. The Journal of biological chemistry 260, 16375–16382 (1985).
Liu, C. C., Kanekiyo, T., Xu, H. & Bu, G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nature reviews Neurology 9, 106–118 (2013).
Huang, Y. et al. Effects of age and amyloid deposition on Abeta dynamics in the human central nervous system. Archives of neurology 69, 51–58 (2012).
Roh, J. H. et al. Disruption of the sleep-wake cycle and diurnal fluctuation of beta-amyloid in mice with Alzheimer’s disease pathology. Science translational medicine 4, 150ra122 (2012).
Acknowledgements
We thank Dr. G. Wu for teaching us to use the JTK_CYCLE algorithm. This research was supported by a grant from the 973 Program (2012CB837702) of the Ministry of Science and Technology of China and by funding from the Beijing Municipal Government. E.E.Z. was supported by the Chinese “Recruitment Program of Global Youth Experts”.
Author information
Authors and Affiliations
Contributions
Z.M. and E.E.Z. conceived the study and designed the experiments. Z.M. and W.J. performed the experiments. Z.M. and E.Z. analyzed the data. Z.M., W.J. and E.E.Z. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Ma, Z., Jiang, W. & Zhang, E. Orexin signaling regulates both the hippocampal clock and the circadian oscillation of Alzheimer’s disease-risk genes. Sci Rep 6, 36035 (2016). https://doi.org/10.1038/srep36035
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep36035
This article is cited by
-
Disturbed circadian rhythm and retinal degeneration in a mouse model of Alzheimer’s disease
Acta Neuropathologica Communications (2023)
-
The circadian clock in the mouse habenula is set by catecholamines
Cell and Tissue Research (2022)
-
Endocytic BDNF secretion regulated by Vamp3 in astrocytes
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.