- Research article
- Published:
The Distributed Annotation System
BMC Bioinformatics volume 2, Article number: 7 (2001)
Abstract
Background
Currently, most genome annotation is curated by centralized groups with limited resources. Efforts to share annotations transparently among multiple groups have not yet been satisfactory.
Results
Here we introduce a concept called the Distributed Annotation System (DAS). DAS allows sequence annotations to be decentralized among multiple third-party annotators and integrated on an as-needed basis by client-side software. The communication between client and servers in DAS is defined by the DAS XML specification. Annotations are displayed in layers, one per server. Any client or server adhering to the DAS XML specification can participate in the system; we describe a simple prototype client and server example.
Conclusions
The DAS specification is being used experimentally by Ensembl, WormBase, and the Berkeley Drosophila Genome Project. Continued success will depend on the readiness of the research community to adopt DAS and provide annotations. All components are freely available from the project website http://www.biodas.org/.
Background
With the rise of computational biology and the decrease in hardware costs, high throughput annotation is now possible within many laboratories. They can now annotate entire genomes relatively quickly and efficiently. What has not kept up with the pace of annotation is the ability for multiple groups to exchange and compare their data, leading to fragmentation of annotation information among multiple databases and web sites, and to a certain level of frustration among the bench biologists who are the intended beneficiaries of this data.
Ideally, an annotation system should give individual experts the ability to contribute to the collective annotation in a quick, robust, and mostly painless fashion. They should have complete control over their annotations in order to keep them current and relevant. These annotations should not need approval from a central authority. Simultaneously, it should be easy for a user to obtain and visualize the most recent data about their particular region of interest. Users would also prefer not to be swamped by bogus information. Unfortunately, these goals seem to be at odds in the current sequence annotation environment.
Initial database efforts were largely centralized repositories such as GenBank, established in 1982 [1]. These databases act primarily as archival storage of sequence information. Consequently, each entry is owned by the sequence provider and integrating annotation information is, by design, nearly impossible.
A number of specialized databases have developed to serve a curatorial role within particular communities, such as Swissprot [2], Refseq [3], and WormPD [4]. A C. elegans database (ACeDB) is one particularly successful community database [5]http://www.acedb.org/. It has served as the central database of phenotyping, bibliographic, mapping, and sequencing information for the Caenorhabditis elegans community since 1990 [6]. Individuals are encouraged to submit annotations and changes to the central database curatorial group. The group then reviews the request and decides what and how it is to be incorporated into the next official release. With limited numbers of curators available, these databases find it difficult to keep up with the requests of many expert annotators.
To overcome the restrictions of archival databases and the bottlenecks of curatorial databases, a number of groups have attempted to develop third party annotation systems. Examples include the Worm Community System [7], the Genome Sequence Database [8], and GDB [9, 10]. These systems typically require global coordination by either keeping all annotations in a centralized open repository or by forcing all parties to adhere to a common database format or by requiring a controlled vocabulary.
Another recent experiment with third party annotation has been the "annotation party," exemplified by Celera's Fly Jamboree and the Human Genome Project Consortium's Analysis Group (HGPCAG). Parties gather together a large number of experts to produce the best annotations possible in a limited time frame. However, it is not clear that the annotation party model is sustainable once the initial flush of enthusiasm has worn off.
The HGPCAG model has a notion of annotation "tracks", where a track contains a particular kind of annotation produced by a particular participating group. For example, the Eddy lab provides a noncoding RNA track that annotates the positions of RNA genes in the human genome. Annotation tracks are independent of each other and therefore easy to integrate into a single display. The concept is essentially identical to the independent columns of annotation displayed by an ACeDB browser, except that the tracks in the HGPCAG annotation are curated by a variety of groups at different institutions, as opposed to a centralized curation group. However, the data for every track are still kept on a single centralized server; updating an annotation track after it has been submitted is cumbersome.
Here we introduce a genome annotation strategy that enables third-party annotation in a way that allows annotators to control and update their work, and which does not require much centralized coordination. The Distributed Annotation System (DAS) was designed as a lightweight system for integrating data from a number of heterogenous distributed databases. The DAS system has a notion of annotation "layers", which are essentially identical to tracks, except that now the data for each layer are on "third party servers" that are controlled by each annotation provider. The key idea was to produce a data exchange standard (the DAS XML specification) that enables layers to be provided in real time from 3rd party servers and overlaid to produce a single integrated view by a DAS client.
Figure 1 shows a cartoon example of the DAS paradigm. The client selects a single reference genome server and any number of annotation servers. The display layers the data returned from each server. A particular annotation can then be queried to retrieve more information from its providing server, as HTML pages.
Basic distributed annotation system architecture One server is the designated reference server, in this case the Washington University Genome Sequencing Center. One or more annotation servers, shown above as Ensembl, Whitehead, and the Sean Eddy Laboratory, provide annotations relative to the reference sequence. The client, at Cold Spring Harbor Laboratory in our example, fetches data from multiple servers and automatically generates an integrated view.
Implementation
The basic system is composed of a genome server, one or more annotation servers, and an annotation viewer. The genome server is responsible for serving genome maps, sequences, and information related to the sequencing process. Annotation servers are responsible for responding to requests on a region and delivering annotations. The client, an annotation viewer, is a lightweight application whose behavior is analogous to a web browser. The viewer communicates with the genome and annotation servers using a well defined language specification.
At a fundamental level, all annotations can be reduced to their coordinates relative to a particular sequence landmark. The DAS viewer retrieves annotations from the various annotation servers and uses the sequence coordinates to generate an integrated index of what is on the genome. This integration is then presented to the user in tabular or graphical form. Annotation providers can provide a suggestion of how their annotations should be rendered in a graphical display, and can provide links back to their databases and web sites to allow the researcher to retrieve further information about the annotation.
Because it relies entirely on sequence coordinates to achieve integration, DAS does not attempt to resolve semantic contradictions between different data sources. The goal of the system is to provide indexing and visualization, thereby making contradictions between annotations visible.
Reference sequence
The distributed annotation system relies on there being a common "reference sequence" on which to base annotations. The reference server consists of a set of "entry points" into the sequence, and the lengths of each entry point. Entry points will vary from genome to genome. For some genome projects, entry points correspond to entire chromosomes. For others, entry points may be a series of contigs.
The entry points describe the top level items on the reference sequence map. It is possible for each entry point to have substructure, basically a series of subsequences (components) and their start and end points. This structure is recursive. Annotations take the form of a statement about a region of the reference sequence. Each annotation is unambiguously located by providing its position as the start and stop positions relative to a "reference sequence."
To give a concrete example, the C. elegans reference map consists of six top level entry points, one per chromosome. Each chromosome is formed from several contigs called "superlinks," and each superlink contains one or more smaller contigs called "links." Links in turn are composed of one or more fully-sequenced clones [11]. One could refer to an annotation by specifying its start or stop positions in clone, link, superlink, or chromosome coordinates.
The reference sequence server is responsible for providing the reference sequence map and the underlying DNA. The server can provide a list of sequence entry points or given a component of the map it can return its parent and children components. The reference server can provide arbitrarily long stretches of raw DNA sequence given a reference subsequence, start position, and stop position. Needless to say, bandwidth becomes a limiting factor for retrieving multi-megabase segments of DNA. However, in practice it is rare for users to retrieve more than a gene's worth of raw DNA at a time.
Annotation servers
Annotation servers are specialized for returning lists of annotations across defined regions of the genome. Each annotation is anchored to the genome map by way of a start and stop position relative to one of the entry points. Annotations have an identifier that is unique to the providing server and a structured description of its nature and attributes. The general description of an annotation follows loosely the general feature format (GFF) which intentionally aims for a basic lowest common denominator description http://www.sanger.ac.uk/Software/formats/GFF/. Annotations may also be associated with URLs where additional human or machine readable information about the annotation can be found.
The annotator is free to describe his annotations using any terms which he feels are appropriate, as DAS does not impose a controlled vocabulary. Annotations have categories, types, and methods defined by the annotator. The annotation type corresponds to a biologically significance description. In the Eddy Lab RNA track of the HGP three types are defined, "tRNA", "snoRNA", and "miscRNA". The annotation method is intended to describe how the annotated feature was discovered, and may include a reference to a software program. The annotation category is a broad functional category. "Homology", "variation" and "transcribed" are example categories. This structure allows researchers to add new annotation types if the existing list is inadequate without entirely losing all semantic value. It is intended that larger annotation servers provide URLs to human-readable information that describes its types, methods and categories in more detail.
Another optional feature of annotation servers is the ability to provide hints to clients on how the annotations should be rendered visually. This is done by returning a DAS "stylesheet." Stylesheets use the type and category information to associate each annotation with a particular graphical representation, a glyph.
Although the servers are conceptually divided between reference servers and annotation servers, there is in fact no key difference between them. A single server can provide both reference sequence information and annotation information. The main functional difference is that the reference sequence server is required to serve the coordinate map and the raw DNA, while annotation servers have no such requirement.
Specification
The main component of DAS is the XML specification, which defines all valid DAS communication. As with HTML, our goal is a language which is human readable, easily parsed, and extensible. The additional file [appendix.pdf] provides a summary of version 1.01 of the DAS specification.
While a client can query multiple servers simultaneously, the communication between the client and any single server follows a simple client server model. Clients query the reference and annotation servers by sending a formatted URL request to each server. Each URL has a site-specific prefix, followed by a standardized path and query string. The standardized path begins with the string /das. This is followed by URL components containing the data source name and a command. For example:
http://stein.cshl.org/das/elegans/features?segment=ZK154:1000,2000
In this case, the site-specific prefix is http://stein.cshl.org/. The request begins with the standardized path /das, and the data source, in this case /elegans. This is followed by the command /features, which requests a list of features relative to a given set of named arguments (?segment=ZK154:1000,2000). The data source component allows a single server to provide information on several genomes.
Servers process the request and return a response as defined by the DAS specification, typically a formatted XML document. The response from the server to the client consists of a standard HTTP header with DAS status information within that header followed optionally by an XML file that contains the answer to the query. The DAS status portion of the header consists of two lines. The first is X-DAS-Version and gives the current protocol version number, currently DAS/1.0. The second line is X-DAS-Status and contains a three digit status code which indicates the outcome of the request. The defined status codes are listed in Table 1.
An example HTTP header: (provided by server)
HTTP/1.1 200 OK
Date: Sun, 12 Mar 2000 16:13:51 GMT
Server: Apache/1.3.6 (Unix) mod_perl/1.19
Last-Modified: Fri, 18 Feb 2000 20:57:52 GMT
Connection: close
Content-Type: text/plain
X-DAS-Version: DAS/1.0
X-DAS-Status: 200
DATA FOLLOWS ...
The specification outlines seven basic queries which a client can use to interrogate a DAS server. The valid queries are briefly summarized in Table 2. Two queries, "dsn" and "entry points", essentially provide information to the client about the structure of the server and the reference sequence. The "dna" query can be used to fetch a segment of DNA from a reference server. A client can request annotations, "features", or a summary of the annotations available, "types", from any DAS server. The main annotation content query, "features", basically follows the general feature format (GFF). The servers provide a "stylesheet" to suggest representations to the client's graphical display. When more information is desired about a particular annotation, the client makes a "link" request. The "link" request, the only query which does not return a structured XML document, returns HTML. It is anticipated that DAS clients will hand off the link requests to the local web browser or other web-accessible genome database.
Prototypes
A series of prototypes for both the client and server components were developed to test various versions of the DAS specification.
Servers
A server is expected to respond to the DAS specification's defined queries with the appropriate content, usually XML. The details of server implementation are left to the various annotation source providers. We provide a sample Perl script for converting ACeDB-based databases into DAS servers, and the Dazzle Java library does the same thing for annotation databases based on the Ensembl code base (T. Down, personal communication, 2001).
The first reference DAS server was written for WormBase [11] and piggybacks on the WormBase software architecture: an Apache/mod_perl web server communicating with an ACeDB database via the AcePerl database access library. The Perl DAS server accepts incoming DAS requests, translates them into the ACeDB query language, reformats the results as XML, and returns them. The WormBase DAS server is currently serving as the C. elegans reference server at http://www.wormbase.org/db/das/. A set of servers containing test data, one reference and four annotation, are available at http://skynet.wustl.edu/cgi-bin/das/.
Viewers
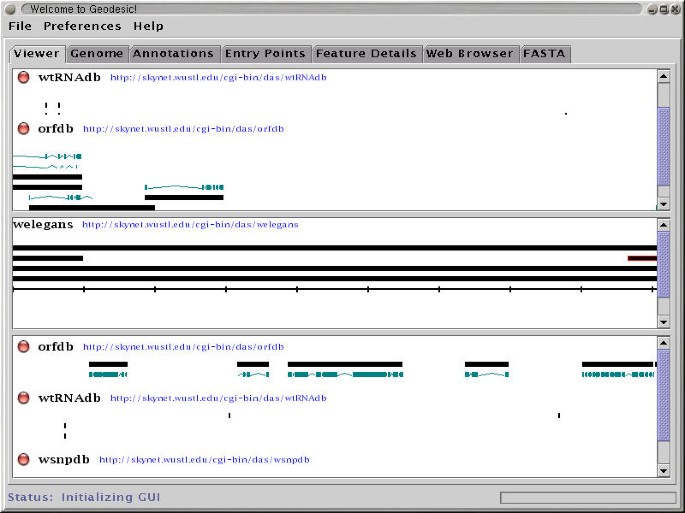
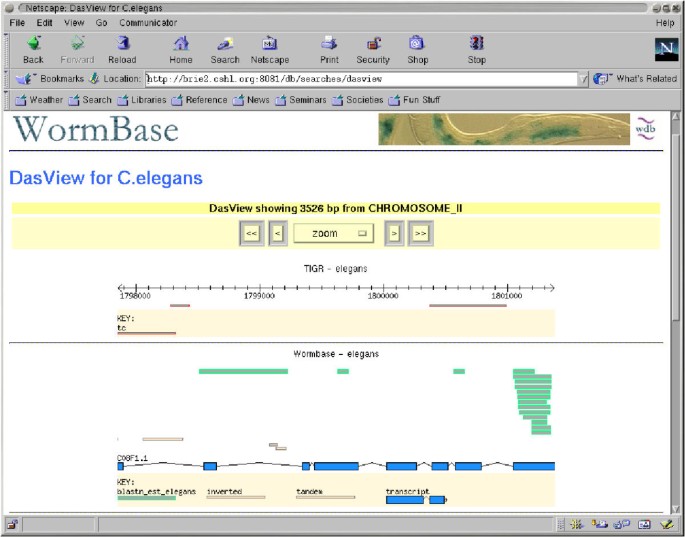
We have developed two prototype DAS client programs. One, called Geodesic, is a stand alone Java application. It connects to one or more DAS servers, retrieves annotations, and displays them in an integrated map, as seen in Figure 2. The other, called DasView, is a Perl application that runs as a server-side script. It connects to one or more DAS servers, constructs an integrated image, and serves the image to a web browser as a set of click-able image map, as seen in Figure 3.
Geodesic is mouse and menu driven. The user can choose which data sources to display. The user identifies a segment of the genome to view by browsing through entry points or entering a region name directly. By clicking on a feature, the user obtains additional information in the Feature Details tab and can optionally follow available links back to the original data source. The user can save displayed data as FASTA, GFF, or DAS XML. The user can, to a limited extent, customize the display within the preferences menu.
The DasView prototype implements an alternative mode of using DAS, browserless server side integration. A database can hook into trusted third party servers behind the scenes. The third party data are then integrated into the normal data displays of the database. In this scenario, no DAS client software would be needed.
Both viewers provide the user with one-click linking back the primary data sources where they can learn more about a selected annotation, and are sufficiently flexible to accept a wide range of annotation types and visualization styles. The stand alone Java viewer is appropriate for extensive, long-term use. The Perl implementation is suitable for casual use because it does not require the user to preinstall the software.
Discussion
DAS distributes data sources across the Internet improving scalability over monolithic systems. This distribution of data encourages a divide-and-conquer approach to annotation, where experts provide and maintain their own annotations. It also permits annotation providers to disagree about a particular region, encouraging informative dissension and dialogue. The separation of sequence and map information from annotation allows them to be stored and represented in a variety of database schema. A number of different database backend alternatives could arise.
The use of links as a method of referencing back to the data provider's web pages provides even greater power of expression and content control. Annotation providers can make available complex query mechanisms for fine access to more information about the data provided to DAS. Alternatively they can link directly to webpages.
DAS does not enforce third party annotations to be peer reviewed. A strict requirement of peer review would block data sharing activities between collaborating labs. However, nothing prevents DAS layers from being "blessed" by a data provider, peer reviewer, or by both.
We made a design decision to use an XML-based format. This gives us a strongly typed, extensible data exchange format, but at the cost of non-trivial bandwidth demands. Bandwidth requirements are a substantial concern in the continued design and development of DAS. A user browsing a large genome can easily request more information than their network connection can reasonably handle. The DAS spec attempts to minimize bandwidth demands by representing each annotation with the minimal set of attributes needed for integration. Further bandwidth reductions will be useful, and the extreme redundancy of XML suggests that compression methods are a natural way forward. The HTTP protocol allows web clients to request byte-level compression of the response by sending the HTTP header "accept-encoding". Web servers can reply with a "content-transfer-encoding" header and a compressed body. The Dazzle server and Bio::Das client have already utilized this feature to reduce their bandwidth requirements. Other compression schema are possible including DAS specific approaches that take advantage of the structure of DAS data.
The World Wide Web Consortium has developed a number of technologies to support XML based systems. A number of these technologies should be considered for future integration into DAS. The Simple Object Access Protocol (SOAP) 1.1 describes a lightweight protocol for the exchange of information in a decentralized, distributed environment. A DAS request may be replaced with a SOAP-style XML-encapsulated document in future versions of this specification. Each annotation is identified by its site-specific database identifier. The combination of this identifier with the server URL and data source produces an feature identifier which is globally unique. Future versions of DAS could utilize this identifier with XPATH and XLINK technologies to permit meta-annotations.
In large part, the continued success of this project will depend on the readiness with which the research community creates annotation sources. To facilitate this, we are working with the BioPerl and BioJava software developer communities http://open-bio.org/ to develop a core set of servers, clients and software modules to support DAS. It is particularly important that the general biological community should be enabled to develop their own DAS annotation servers, without learning XML and Web software development. Easy, well-documented DAS annotation servers that take input data in simple flat file formats and convert it automatically to DAS XML are currently under development.
The DAS specification is under continued development. It does not detail how data source URLs will be publicized. It is anticipated that word of mouth and publications will be the driving forces in user selection. In addition, search engines can be developed to work with the DAS specification.
Conclusions
The DAS specification is already being used in real-world applications. The July 9 2001 release of the Ensembl database of human genome annotations contains support for DAS, including an integrated DAS viewer and multiple annotation servers (M. Pocock, personal communication, 2001). The WormBase DAS server has recently been supplemented by a third party annotation source of cDNA alignments contributed by The Institute for Genome Research, and a prototype DAS reference server for the Drosophila genome is also available, courtesy of the Berkeley Drosophila Genome Project (B. Marshall, personal communication, 2001). Table 3 lists the URLs where one can learn more about the current state of the art in DAS implementations.
References
Smith TF: The history of genetic sequence databases. Genomics 1990, 6: 701–707.
Bairoch A, Apweiler R: The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Research 1999, 27: 49–54. 10.1093/nar/27.1.49
Pruitt KD, Katz KS, Sicotte H, Maglott DR: Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet 2000, 16: 44–47. 10.1016/S0168-9525(99)01882-X
Costanzo MC, Crawford ME, Hirschman JE, Kranz JE, Olsen P, Robertson LS, Skrzypek MS, Braun BR, Hopkins KL, Kondu P, et al.: YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res 2001, 29: 75–9. 10.1093/nar/29.1.75
Eeckman FH, Durbin R: ACeDB and macace. Methods Cell Biol 1995, 48: 583–605.
Waterson R, Sulston J: The genome of the Caenorhabditis elegans. Proc. Natl. Acad. Sci 1995, 92: 10836–10840.
Shoman LM, Grossman E, Powell K, Jamison C, Schatz BR: The Worm Community System, release 2.0 (WCSr2). Methods Cell Biol 1995, 4: 607–625.
Skupski MP, Booker M, Farmer A, Harpold M, Huang W, Inman J, Kiphart D, Root S, Schilkey F, Schwertfeger J, et al.: The Genome Sequence DataBase: towards an integrated functional genomics resource. Nucleic Acids Res 1999, 27: 35–38. 10.1093/nar/27.1.35
Letovsky SI, Cottingham RW, Porter CJ, Li PW: GDB: the human genome database. Nucleic Acids Res 1998, 26: 94–99. 10.1093/nar/26.1.94
Cuticchia AJ: Future vision of the GDB human genome database. Hum Mutat 2000, 15: 62–67. 10.1002/(SICI)1098-1004(200001)15:1<62::AID-HUMU13>3.0.CO;2-R
Stein L, Sternberg P, Durbin R, Thierry Mieg J, Spieth J: WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res 2001, 29: 82–86. 10.1093/nar/29.1.82
Acknowledgements
The initial ideas for DAS were developed in conversations with LaDeana Hillier of the Washington University Genome Sequencing Center. This work was primarily supported by NIH National Human Genome Research Institute (NHGRI) grant 2-P01-HG00956 for the Caenorhabditis elegans genome project, and by a Howard Hughes Medical Insitute (HHMI) Predoctoral Fellowship to RDD. We also gratefully acknowledge additional funding support from HHMI and the NIH NHGRI.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
About this article
Cite this article
Dowell, R.D., Jokerst, R.M., Day, A. et al. The Distributed Annotation System. BMC Bioinformatics 2, 7 (2001). https://doi.org/10.1186/1471-2105-2-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2105-2-7