Abstract
The involvement of Amyloid-β (Aβ) in the pathogenesis of Alzheimer’s disease (AD) is well established. However, it is becoming clear that the amyloid load in AD brains consists of a heterogeneous mixture of Aβ peptides, implying that a thorough understanding of their respective role and toxicity is crucial for the development of efficient treatments. Besides the well-studied Aβ40 and Aβ42 species, recent data have raised the possibility that Aβ43 peptides might be instrumental in AD pathogenesis, because they are frequently observed in both dense and diffuse amyloid plaques from human AD brains and are highly amyloidogenic in vitro. However, whether Aβ43 is toxic in vivo is currently unclear. Using Drosophila transgenic models of amyloid pathology, we show that Aβ43 peptides are mainly insoluble and highly toxic in vivo, leading to the progressive loss of photoreceptor neurons, altered locomotion and decreased lifespan when expressed in the adult fly nervous system. In addition, we demonstrate that Aβ43 species are able to trigger the aggregation of the typically soluble and non-toxic Aβ40, leading to synergistic toxic effects on fly lifespan and climbing ability, further suggesting that Aβ43 peptides could act as a nucleating factor in AD brains. Altogether, our study demonstrates high pathogenicity of Aβ43 species in vivo and supports the idea that Aβ43 contributes to the pathological events leading to neurodegeneration in AD.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is a devastating neurodegenerative disorder characterized by the presence of two neuropathological hallmarks, namely the intraneuronal deposition of hyperphosphorylated Tau proteins into neurofibrillary tangles and accumulation of Aβ peptides both intracellularly and into extracellular amyloid plaques. Aβ peptides are produced following the sequential proteolytic cleavage of their precursor protein, APP, by secretases. The cleavage releasing the C-terminal part of Aβ can occur at different residues and hence produce peptides of different lengths, ranging from 37 to 49 amino acids [2], among which Aβ40 and Aβ42 are the most abundant [32]. Aβ40 species are soluble and abundantly produced in both healthy and AD brains. In contrast, Aβ42 levels are substantially increased in AD brains. Because of their high propensity to aggregate due to their two additional hydrophobic residues, Aβ42 peptides are the main constituents of amyloid deposits [36] and many studies have shown that they are highly pathogenic in the context of AD [15, 37].
Interestingly, recent studies have pointed to the potential of other Aβ species, and in particular of Aβ43, to be involved in AD pathogenesis. Indeed, Aβ43 is significantly increased in AD brains, deposits more frequently than Aβ40 and is found in the core of amyloid plaques [13, 17, 27, 30, 36]. Moreover, recent data suggest that Aβ43 is highly amyloidogenic in vitro [3, 4, 15, 29] and reduces the viability of cultured neuronal cells when applied in the culture medium [1, 23, 29]. In addition, higher cortical Aβ43 levels have been associated with increased amyloid load and impaired memory in the APP/PS1-R278I transgenic mouse model [29].
Importantly, in addition to its ability to self-aggregate in vitro to induce neurotoxicity, Aβ43 has been suggested to initiate the seeding of other Aβ peptides. Its addition to a mixture of Aβ peptides was shown to accelerate the formation of Thioflavin T-positive amyloid structures in vitro, in a more potent manner than did Aβ42 or Aβ40 [29]. In addition, Aβ43 was shown to deposit earlier than other Aβ species in the brain of mouse models of AD [38] and to be surrounded by other Aβ species in brains of AD patients [29], further suggesting its ability to nucleate and subsequently titrate other Aβ species.
However, a direct in vivo demonstration that Aβ43 self-aggregates, triggers neurotoxicity and exacerbates neurotoxicity from other Aβ species is so far lacking. The fruit fly Drosophila has proved an excellent in vivo model system for the analysis of both loss of function [10, 25] and toxic gain of function [5, 24] human neurodegenerative diseases. We have thus generated inducible transgenic Drosophila lines expressing human Aβ43, Aβ42 or Aβ40, using an attP/attB site-directed integration strategy to ensure both standard levels of mRNA expression and the best ratio of induced versus basal expression [20]. We observed that Aβ43 was highly insoluble in vivo and that it led to severe toxic effects, both when constitutively expressed in the compound eye of the fly, leading to eye roughness, and when specifically induced in the adult nervous system, as measured by a progressive loss of photoreceptor neurons, impaired locomotion and decreased lifespan. Interestingly, by combining transgenes encoding different Aβ isoforms we also found that, in presence of Aβ43, Aβ40 species were progressively shifted from the soluble to the insoluble protein fraction and that the overall Aβ insolubility was increased, leading to significant defects in climbing ability and survival. Altogether, our results demonstrate high pathogenicity of Aβ43 species in vivo and delineate their ability to trigger toxicity from the otherwise innocuous Aβ40.
Materials and methods
Generation of transgenic fly lines
Human Aβ1–40, Aβ1–42 and Aβ1–43 bearing a secretion signal from the Drosophila necrotic gene [6] were synthesized by MWG Operon (Germany) using insect optimized codons and were subsequently cloned into the pUASTattB vector. An identical cDNA was used for all sequences that were common to all lines. Transgenic fly lines were generated using the φC31 and attP/attB targeted integration system and transgenes were inserted into the attP40 or attP2 landing-site loci to ensure both standard levels of mRNA expression and the best ratio of induced versus basal expression [20]. Transgenic fly lines were verified by sequencing of the corresponding genomic DNA and by western blotting (Fig. 1b). The gene-switch Elav-Gal4 inducible line (elavGS) was derived from the original elavGS 301.2 line [26] and obtained as a generous gift from Dr. H. Tricoire (CNRS, France). The GMR-Gal4 line was obtained from the Bloomington Drosophila Stock Center.
Expression of Aβ in fly heads. a qRT-PCR analysis of Aβ mRNA levels measured in head extracts of Aβ40, Aβ42 and Aβ43 transgenic lines (p > 0.05, one-way ANOVA) following expression in the fly nervous system using the neuron-specific elavGS driver. b Western blot analysis of head protein extracts from Aβ40, Aβ42 and Aβ43 transgenic fly lines (elavGS driver) probed with Aβ40-, Aβ42-, and Aβ43-specific antibodies, showing the high specificity of the transgenic lines
Fly maintenance
All fly stocks were kept at 25 or 29 °C on a 12:12 h light:dark cycle at constant humidity and fed with standard sugar/yeast/agar (SYA) medium (15 g L−1 agar, 50 g L−1 sugar, 100 g L−1 yeast, 30 mL L−1 nipagin and 3 mL L−1 propionic acid). All lines were backcrossed into a white Dahomey (wDah) wild-type outbred strain for at least ten generations prior to experiments. Adult-onset transgene expression was achieved using the inducible gene-switch UAS-Gal4 system and through addition of the activator RU486 (Mifepristone) to fly food at a final concentration of 200 µM. Non-induced controls were obtained by adding the vehicle (i.e. ethanol) to fly food. All experimental flies were kept at 25 °C throughout development and during the 48-h mating step following eclosion, after which females were sorted and transferred to 29 °C. Only flies used for the eye phenotype experiment were kept at 25 °C, since an eye phenotype can already be observed in the GMR-Gal4 driver control at 29 °C.
All investigated lines were homozygous for the Aβ transgene unless indicated by “1×Aβ”, meaning flies were heterozygous for the transgene.
Lifespan analysis
For lifespan experiments, 200 once-mated females per group were allocated to vials at a density of 10 flies per vial and subsequently kept at 29 °C. Flies were transferred to new vials every 2–3 days and the number of dead flies was recorded. Lifespan results are expressed as the proportion of survivors ±95 % confidence interval.
Climbing assay
Climbing assays were performed blindly using a countercurrent apparatus as previously described [9] using at least 3 replicates of 20 female flies per group. One hour prior to the measurement, flies were randomized and transferred to plastic tubes for acclimation. Flies were placed into the first chamber of the six-compartment climbing apparatus, tapped to the bottom and given 20 s to climb a distance of 15 cm, after which flies above this level were shifted to the second chamber. Both sets of flies were tapped again to the bottom and allowed to climb for another 20 s. This procedure was repeated for a total of 1 min and 40 s so that flies could climb into the six chambers, and the number of flies in each chamber was counted at the end of the experiment. The climbing index (CI) was calculated as previously described [9] and varied between 0 (all flies stayed in the first compartment) and 1 (all flies reached the last chamber).
Eye phenotype
Eye images of 6-day-old female flies expressing Aβ under the control of the GMR-Gal4 driver at 25 °C were taken using a Leica M165 FC stereo microscope. At least 8 flies per genotype were investigated.
Rhabdomere assay
We used the cornea neutralization technique [8] to visualize the rhabdomeres from the ommatidia of the fly compound eye. Briefly, dissected fly heads were mounted on a microscope slide using a drop of nail polish and further covered with oil. The number of ommatidia lacking rhabdomeres was counted using a Leica DMI4000B/DFC 340FX inverted microscope and a 40× oil immersion objective. At least 50 ommatidia per fly and 5 flies per genotype were examined.
Total Aβ levels
20 to 25 female heads per biological replicate were homogenized in 100 µL of 70 % formic acid using a disposable pellet mixer and a plastic Eppendorf pestle. Samples were centrifuged at 16,000g for 20 min at room temperature. The supernatant was collected and subsequently evaporated using a SpeedVac. The dry pellet was resuspended in 100 µL 2× LDS containing reducing agent (Invitrogen) and homogenized by sonication (10 pulses). Samples were then boiled at 100 °C for 10 min and 10 µL of each sample were used for western blotting to determine total Aβ levels.
Fractionation of soluble and insoluble Aβ peptides
The procedure was based on previous reports [7] with some modifications. Briefly, 20 female heads per biological replicate were homogenized in 100 µL ice-cold RIPA buffer (Pierce) supplemented with SDS at a final concentration of 1 % and Complete mini without EDTA protease inhibitor (Roche) using a disposable pellet mixer and a plastic Eppendorf pestle. Samples were incubated on ice for 30 min and were then centrifuged at 100,000g for 1 h at 4 °C in an Optima XPN-100 ultracentrifuge (Beckman Coulter). The supernatant (“soluble fraction”) was collected and the pellet was homogenized in 100 µL of 70 % formic acid by pipetting, followed by 5-min incubation in a sonication bath. Samples were centrifuged again at 100,000g for 1 h at 4 °C, after which the supernatant was collected (“insoluble fraction”) and evaporated using a SpeedVac. The dry pellet was then resuspended in 100 µL 2× LDS containing reducing agent (Invitrogen) by pipetting followed by 5-min incubation in a sonication bath. Protein concentration of the soluble fraction was measured using the BCA protein assay kit (Pierce), and 30 µg of soluble proteins and equivalent volumes of insoluble proteins were used for western blotting. Results are represented as the proportion of insoluble Aβ species, i.e. insoluble Aβ/(soluble Aβ + insoluble Aβ) × 100. Results are expressed as mean ± sem.
Sample preparation for soluble Aβ oligomers and dot blotting
The procedure was based on previous reports [21] with some modifications. Briefly, 20 female heads per biological replicate were homogenized in 100 µL ice-cold 1× PBS buffer supplemented with Complete mini without EDTA protease inhibitor (Roche) using a disposable pellet mixer and a plastic Eppendorf pestle. Samples were then ultra-centrifuged at 78,000g for 1 h at 4 °C. The supernatant, i.e. the PBS-soluble fraction was collected and protein concentration was measured using the BCA protein assay kit (Pierce). One microlitre per sample (corresponding to 1.5 µg of proteins) was spotted onto a 0.2 µm nitrocellulose membrane (GE Healthcare) and let to dry for 30 min. The membrane was then blocked in TBS-low tween buffer (containing 0.01 % Tween) with 10 % non-fat dry milk for 1 h at room temperature, incubated overnight at 4 °C with the A11 anti-oligomer antibody (1/1000, Invitrogen) and then for 1 h at room temperature with HRP-conjugated anti-rabbit antibody (1/10,000, Invitrogen). Detection was performed using ECL prime chemiluminescence kits (GE Healthcare) and Hyperfilms (GE Healthcare).
Sample preparation for LDS/SDS-stable Aβ oligomers
20 female heads per biological replicate were homogenized in 100 µL 2× LDS containing reducing agent (Invitrogen) using a disposable pellet mixer and a plastic Eppendorf pestle. Sample were incubated on ice for 30 min and were then boiled at 100 °C for 10 min. 15 µL per sample were used for western blotting to evaluate LDS/SDS-stable Aβ oligomers.
Western blotting
Protein samples were separated on 16.5 % Tris-Tricine Criterion gels (Biorad) and subsequently transferred to 0.2 µm nitrocellulose membranes (GE Healthcare). After a boiling step of 4 min in 1× PBS, membranes were blocked in TNT buffer (Tris–HCl 15 mM pH 8, NaCl 140 mM, 0.05 % Tween) with 5 % non-fat dry milk for 1 h at room temperature and incubated overnight at 4 °C with the following primary antibodies: anti-Aβ1–16 mAb (6E10, 1/5000, Covance), anti-Aβ1–40 mAb (9682, 1/500, Cell Signaling), anti-Aβ1–42 mAb (12F4, 1/500, Covance), anti-Aβ1–43 mAb (9C4, 1/500, Covance), anti-α-tubulin (11H10, 1/5000, Cell Signaling) and anti-β-actin (1/100,000, Abcam). HRP-conjugated anti-mouse or anti-rabbit antibodies (1/10,000, Invitrogen) were used for 1 h at room temperature and detection was performed using ECL or ECL prime chemiluminescence kits (GE Healthcare) and Hyperfilms (GE Healthcare). Bands were quantified using the ImageJ software (Scion Software) and results are expressed as mean ± sem.
Immunofluorescence
After decapitation and removal of proboscis, heads of 20-day-old GMR-Gal4-driven Aβ flies were fixed for 3 h in 4 % paraformaldehyde in PBS and then incubated overnight in 25 % sucrose in PBS. Heads were frozen in Tissue-Tek O.C.T. (Sakura Finetek) and kept at −80 °C until use. Immunofluorescence was performed on 16 µm cryosections using an anti-Aβ1–40 mAb antibody (D8Q71, 1/200, Cell Signaling) followed by anti-rabbit Alexa-488 secondary antibody (1/250, Invitrogen), both being diluted in PBS with 0.1 % Triton and 5 % non-fat dry milk. An incubation step of 3 min in 70 % formic acid was included prior to blocking to unmask antigens. Stained sections were mounted using Vectashield with DAPI (Vector) and analysed with a Leica DMI4000B/DFC 340FX inverted microscope. Quantification of Aβ40 deposits was performed using ImageJ from at least 6 flies per genotype.
RNA extraction and qRT-PCR
Total RNA was extracted from 25 female heads per replicate using a Trizol-Chloroform-based procedure (Invitrogen) and subsequently treated with DNAse I (Ambion). 300 ng of RNA were then subjected to cDNA synthesis using the SuperScript Vilo Mastermix (Invitrogen). Quantitative real-time PCR was performed using TaqMan primers (Applied Biosystems) in a 7900HT real-time PCR system (Applied Biosystems). RPL32 and actin5c were used as normalization controls and the relative expression of target genes was determined by the ΔΔC T method. Six independent biological replicates per group were analysed. Results are expressed as a percentage of the corresponding control transgenic line and are plotted as mean ± sem.
Statistical analysis
For lifespan experiments, statistical differences were assessed using the log-rank test. Eye phenotypes were statistically evaluated using the Fisher’s exact test. Other results are expressed as mean ± sem and differences between mean values were determined using either Student’s t test, one-way ANOVA followed by Tukey’s post hoc test or two-way ANOVA followed by Tukey’s post hoc test, using Graphpad Prism software. p values <0.05 were considered significant.
Results
We generated transgenic Drosophila lines expressing human Aβ43, Aβ42 or Aβ40, using a site-directed integration strategy to allow transgene insertion into the same genomic locus and therefore ensure equivalent levels of Aβ mRNA expression among the lines, as verified by qRT-PCR (p > 0.05, Fig. 1a). Specific expression of individual Aβ species was checked by western blot using C-terminal-specific, anti-Aβ antibodies (Fig. 1b), confirming that each transgenic line was expressing a single Aβ species.
We first examined the effect of human Aβ homozygous over-expression in Drosophila eyes, using the constitutive, eye-specific GMR-Gal4 driver. While the eyes of Aβ40-expressing flies were not affected and appeared similar to those of the driver line control, Aβ43 expression led to ommatidial disorganization and eye roughening, indicating that Aβ43 was able to induce toxicity in vivo (Fig. 2a). Quantification of the observed rough eye phenotypes among the lines confirmed a significant toxic effect of Aβ43 expression (**p < 0.01, Aβ43 vs. Aβ40 and Aβ43 vs. GMR driver control, Fisher’s exact test, supplementary Fig. 1), although this effect was significantly milder than the one induced by Aβ42 (#p < 0.05 and *p < 0.05, Aβ43 vs. Aβ42, Fisher’s exact test, supplementary Fig. 1). We then used the pan-neuronal and RU486-inducible elav-GeneSwitch-Gal4 (elavGS) driver [28] to investigate the effects on fly climbing ability and survival of the expression of human Aβ specifically in adult neurons. As expected from previous reports [7, 33], adult-restricted, neuronal expression of Aβ42 led to severe toxic effects, both on climbing ability (p < 0.0001 vs. non-induced controls after 11 and 18 days of induction, two-way ANOVA, Fig. 2b) and on fly survival (median lifespan: −52.4 % of non-induced controls, p < 0.0001, log-rank test, Fig. 2c), while induction of Aβ40 produced no overt defects in climbing ability (p > 0.05 vs. non-induced controls at all investigated ages, two-way ANOVA, Fig. 2b) or survival (median lifespan: −5.6 % of non-induced controls, Fig. 2c), similar to what we observed for the driver line control (climbing ability: p > 0.05 at all investigated ages using two-way ANOVA, supplementary Fig. 2a; median lifespan: −6.3 % of non-RU486-fed controls, supplementary Fig. 2b). In line with the toxicity measured following its specific over-expression in the fly eye, inducing Aβ43 expression in the adult nervous system severely impaired fly climbing performance (****p < 0.0001 vs. non-induced controls after 18 days of induction, two-way ANOVA, Fig. 2b), although the induced toxicity was significantly milder than following Aβ42 expression (p < 0.0001 and p < 0.001 after 11 and 18 days of induction, respectively, induced-Aβ42 vs. induced-Aβ43, two-way ANOVA, Fig. 2b). Survival showed a similar pattern, with neuronal expression of Aβ43 leading to a severe shortening of median lifespan, corresponding to −41.6 % of the non-induced controls (p < 0.0001, log-rank test, Fig. 2c). Our results therefore demonstrate strong toxicity from Aβ43 over-expression in adult neurons.
Aβ43 expression led to detrimental effects on fly eye appearance, climbing ability and survival. a Representative microphotographs of adult fly eyes upon Aβ over-expression using the constitutive, eye-specific GMR-Gal4 driver. Control eyes were obtained from the GMR-Gal4 driver line, which do not over-express any Aβ peptide. Climbing ability (b) and survival curves (c) of RU486-induced (plain curves) and non-induced (dotted curves) Aβ40 (blue), Aβ42 (purple) and Aβ43 (red) transgenic flies. The inset shows the percentage of median lifespan reduction versus non-induced control. ****p < 0.0001 versus non-induced controls, two-way ANOVA
To investigate whether neuronal expression of Aβ resulted in neuronal degeneration, we used the quantitative pseudopupil technique to analyse the loss of photoreceptor neurons from the fly compound eye [8, 14, 35]. Each ommatidium from the fly eye contains seven visible photoreceptor neurons that produce photosensitive structures called rhabdomeres. We evaluated the proportion of ommatidia showing a loss of rhabdomeres following Aβ expression in adult neurons (Fig. 3, the bottom inset showing an ommatidium lacking the central rhabdomere, to be compared with a normal ommatidium in Fig. 3, top inset). While expression of Aβ40 led to no significant loss of photoreceptor neurons as compared to the elavGS driver control (p > 0.05 vs. age-matched controls, at all investigated ages, Fig. 3), Aβ42-expressing flies displayed a progressive and pronounced neurodegeneration, the percentage of affected ommatidia reaching 10.48 ± 1.93 % at day 13 (***p < 0.001 vs. age-matched elavGS control, one-way ANOVA, Fig. 3) and 20.02 ± 2.52 % at day 19 (***p < 0.001 vs. age-matched elavGS control, one-way ANOVA, Fig. 3). Interestingly, we found that the expression of Aβ43 in adult neurons also triggered progressive loss of rhabdomeres with age (p < 0.01, one-way ANOVA), affecting 3.18 ± 0.79 and 8.64 ± 2.28 % of ommatidia at day 19 and 25, respectively, the latter being significantly higher than the effect observed in both elavGS controls and Aβ40-expressing flies (*p < 0.05 vs. 25-day-old elavGS driver controls and p < 0.01 vs. 25-day-old Aβ40 transgenic line, one-way ANOVA, Fig. 3), showing that Aβ43 peptides led to neurodegeneration in vivo. This neuronal loss was significantly milder than that observed following Aβ42 over-expression (p < 0.001 at day 13 and day 19, Aβ43 vs. Aβ42, one-way ANOVA; note that all Aβ42-expressing flies were dead at day 25). Altogether, our data demonstrate the ability of Aβ43 peptides to trigger toxicity and neurodegeneration in Drosophila, and suggest that the toxic effects are significantly milder than those induced by Aβ42 over-expression.
Aβ43 expression led to progressive neurodegeneration. The percentage of ommatidia lacking rhabdomeres was quantified from the RU486-induced Aβ40, Aβ42 and Aβ43 transgenic fly lines and elavGS driver control at three different time points [13 (white), 19 (grey) and 25 (black) days of induction, *p < 0.05 and ***p < 0.001 vs. age-matched elavGS driver control]. Insets show representative micrographs of the rhabdomeres from a normal ommatidium (a) and from an ommatidium lacking the central rhabdomere (b). All Aβ42-expressing flies were dead at day 25
Since Aβ mRNA levels were comparable among the lines as measured by quantitative real-time PCR (Fig. 1a), we hypothesized that the different degrees of toxicity induced by over-expression of Aβ40, Aβ42 or Aβ43 could be explained by a differential stability of the Aβs at the peptide level. We therefore measured the total levels of Aβ peptides retrieved from fly heads following Aβ induction in the adult nervous system, using an antibody binding to the N-terminal region of Aβ that is common to all three peptides. First, we analysed total Aβ levels at a very early time point, following 4 h of RU486 induction, and we observed no significant difference between the lines (p > 0.05, one-way ANOVA, Fig. 4a). However, as early as 1 day following the beginning of transgene induction, we could already observe that total Aβ amounts significantly differed between the lines, with levels of Aβ40 and Aβ43 representing 55.92 ± 3.45 and 79.30 ± 2.55 % of Aβ42 levels, respectively (*p < 0.05 and ***p < 0.001, one-way ANOVA, Fig. 4b). Such differences intensified with time. Total Aβ peptide levels measured in the Aβ40-expressing line were extremely low compared to Aβ42 transgenics after 5 days of induction (2.79 ± 1.32 % relative to Aβ42 levels, ***p < 0.001, one-way ANOVA, Fig. 4c), in line with previous reports [7]. We also observed that total levels of Aβ were markedly lower in the Aβ43 than in the Aβ42 transgenics after 5 days of induction (33.60 ± 7.96 % relative to Aβ42 levels, ***p < 0.001, one-way ANOVA, Fig. 4c), but significantly higher than those retrieved from the heads of Aβ40 transgenic flies (*p < 0.05, one-way ANOVA, Fig. 4c).
Total Aβ amounts and solubility following induction in the fly nervous system. Western blot analysis of total Aβ retrieved from fly head extracts following either 4 h (a), 1 day (b) or 5 days (c) of neuronal (elavGS-driven) expression of Aβ40, Aβ42 or Aβ43, using the pan-Aβ 6E10 antibody. Quantifications (right panels) were made using Tubulin for normalization and results are expressed relative to levels measured in the Aβ42 line (4 h induction: p > 0.05; 1 day induction: *p < 0.05, Aβ43 vs. Aβ42 and Aβ43 vs. Aβ40, ***p < 0.001, Aβ42 vs. Aβ40; 5 days induction: *p < 0.05, Aβ40 vs. Aβ43, ***p < 0.001, Aβ42 vs. Aβ40 and Aβ42 vs. Aβ43). d Fractionation of SDS-soluble (S) and SDS-insoluble/formic acid-soluble (I) Aβ species was performed from heads of elavGS-driven Aβ40, Aβ42 and Aβ43 transgenic flies following 1 or 5 days of induced expression and probed using the 6E10 pan-Aβ antibody. The quantifications of the proportion of insoluble Aβ species are shown on the right panel (***p < 0.001, Aβ42 vs. Aβ43 and ****p < 0.0001, Aβ40 vs. Aβ42 and Aβ40 vs. Aβ43 after 1 day of induced expression; *p < 0.05, Aβ42 vs. Aβ43; ***p < 0.001, Aβ40 vs. Aβ42 and Aβ40 vs. Aβ43 after 5 days of induced expression)
These results suggest that these Aβ isoforms differentially accumulate in the fly nervous system. Such effects could be explained by a differential aggregation propensity and therefore protein stability of these three Aβ species. To tackle this question, we performed fractionation experiments to evaluate whether Aβ40, Aβ42 and Aβ43 would display an unequal propensity to form amyloid structures in vivo in the adult fly brain, as previously suggested by in vitro studies [4, 29]. Interestingly, fractionation of Aβ peptides according to their solubility in 1 % SDS revealed differences among the lines as early as 1 day following the start of induction in the fly nervous system. We confirmed previous reports from Drosophila [7, 12] of a high solubility of Aβ40 species (0.48 ± 0.10 and 1.61 ± 0.66 % of Aβ40 species being insoluble after 1 and 5 days of induction, respectively, Fig. 4d) while most of the Aβ42 was found in the insoluble fraction (51.92 ± 2.94 % at day 1 and 94.76 ± 1.61 % at day 5, ****p < 0.0001 vs. Aβ40, one-way ANOVA, Fig. 4d), suggesting that Aβ42 peptides form insoluble structures when expressed in adult fly neurons. Interestingly, 1 day after the beginning of induction, a significantly higher proportion of Aβ43 peptides than Aβ40 were found in an insoluble state (30.30 ± 1.50 %, ****p < 0.0001 vs. Aβ40, one-way ANOVA, Fig. 4d), although they were more soluble than Aβ42 peptides (***p < 0.001 vs. Aβ42, one-way ANOVA, Fig. 4d). The proportion of insoluble Aβ43 rose to 89.17 ± 0.99 % after 5 days of induction (****p < 0.0001 vs. Aβ40, one-way ANOVA, Fig. 4d), indicating that Aβ43 peptides were mostly insoluble at this stage. However, Aβ43 was still significantly more soluble than Aβ42 (*p < 0.05, Aβ43 vs. Aβ42, one-way ANOVA, Fig. 4d).
Such drastic differences in Aβ solubility might reflect differential ability of cells to clear Aβ peptides. To test this hypothesis, we performed a “switch-on/switch-off” experiment, in which flies were first exposed to the RU486 inducer for a period of 5 days (“5d ON”), to activate neuronal Aβ expression, and then transferred to RU486-free food for either 2 or 7 days (“5d ON + 2d OFF” and “5d ON + 7d OFF”, respectively) to evaluate Aβ clearance either shortly following induction arrest or at a later stage to ensure the complete removal of RU486 from the fly nervous system [26, 28]. A parallel cohort was exposed to the inducer for the entire duration of the experiment, i.e. 12 days (“12d ON”). Using the 6E10 pan-Aβ antibody, we evaluated the total levels of Aβ retrieved from fly head extracts under these conditions (Fig. 5). Interestingly, clearance of Aβ appeared strikingly different depending on the investigated isoform. While Aβ40 was rapidly cleared from the fly nervous system, with 19.26 ± 3.03 and 0.36 ± 0.04 % of Aβ40 remaining 2 and 7 days following induction arrest, respectively (relative to the levels measured directly following 5 days of induction; ****p < 0.0001, 5d ON + 2d OFF vs. 5d ON and **p < 0.01, 5d ON + 7d OFF vs. 5d ON + 2d OFF, one-way ANOVA, Fig. 5a, b), we could not observe any reduction in Aβ42 levels 2 days following induction arrest (Fig. 5a). Rather, Aβ42 appeared to accumulate in the fly nervous system, reaching 349.25 ± 50.77 % of the levels observed before the switch to RU486-free conditions (**p < 0.01, 5d ON + 2d OFF vs. 5d ON, one-way ANOVA, Fig. 5b). Seven days following induction arrest, Aβ42 levels represented 209.91 ± 33.60 % of the initial “5d ON” levels, suggesting that the clearance process of accumulated Aβ42 was effective at this stage (*p < 0.05, 5d ON + 7d OFF vs. 5d ON + 2d OFF, one-way ANOVA, Fig. 5a, b), although the retrieved Aβ42 amounts did not reach lower levels than those measured in the “5d ON” condition (p > 0.05, 5d ON + 7d OFF vs. 5d ON, one-way ANOVA, Fig. 5b). Interestingly, the behaviour of Aβ43 following induction arrest appeared strikingly different from that of both Aβ40 and Aβ42. Whereas 2 days following induction arrest there was a trend for reduced Aβ43 amounts (40.18 ± 9.90 % vs. 5d ON, p > 0.05 using one-way ANOVA, Fig. 5a, b), we could not observe any further change in Aβ43 levels following 7 days of induction arrest (p > 0.05, 5d ON + 7d OFF vs. 5d ON + 2d OFF, one-way ANOVA, Fig. 5a, b). Additionally, the analysis of Aβ43 amounts retrieved from head extracts following 12 days of RU486-induced expression revealed a significant Aβ43 accumulation in the fly nervous system upon induction (248.09 ± 20.84 % at 12d ON relative to 5d ON levels, **p < 0.01, one-way ANOVA, Fig. 5a, b), while Aβ40 expression appeared to yield rather stable total Aβ levels over time (127.27 ± 19.09 % at 12d ON, p > 0.05 vs. 5d ON, one-way ANOVA, Fig. 5a, b). In contrast, Aβ42 accumulation was striking, reaching after 12 days of induction 525.70 ± 46.10 % of the levels observed following 5 days of induction (***p < 0.001, one-way ANOVA, Fig. 5a, b). Altogether, these results suggest on one hand that Aβ43 peptides accumulate in the fly nervous system, though to a lower extent than Aβ42, and on the other hand that cells appear to be less efficient in the clearance of Aβ43 as compared to that of Aβ40. Together with the phenotypic effects of these Aβ isoforms (Figs. 2, 3), these results suggest that Aβ43-induced toxicity is related to its high propensity to self-aggregate and accumulate in vivo.
Clearance of Aβ peptides following induction arrest. a Western blot analysis of total Aβ amounts retrieved from adult fly head extracts following neuronal (elavGS-driven) expression of Aβ40 (top panel), Aβ42 (middle panel) or Aβ43 (lower panel) following either 5 days of RU486 induction (“5d ON”), 5 days of RU486 induction followed by exposure to RU486-free food for either 2 or 7 days (“5d ON + 2d OFF” and “5d ON + 7d OFF”, respectively), or 12 days of RU486 induction (“12d ON”). Aβ detection was performed using the pan-Aβ 6E10 antibody and Tubulin levels were used for normalization. b Quantification of total Aβ levels from a. Results were normalized to Tubulin and are expressed relative to levels measured in the “5d ON” condition for each genotype (*p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 using one-way ANOVA followed by Tukey’s post hoc test)
As increasing evidence suggest that toxic Aβ oligomers are important effectors of neurodegeneration (for review, see [2]), we evaluated the presence of PBS-soluble Aβ oligomeric species in our transgenic Aβ fly lines by dot blotting (supplementary Fig. 3a). Immuno-labelling of the membrane with the A11 anti-oligomer antibody did not highlight any specific signal corresponding to soluble oligomers in our transgenic Aβ lines as compared to the elavGS driver control (p > 0.05, one-way ANOVA, supplementary Fig. 3a). However, since this antibody is not expected to detect low-molecular weight (MW) Aβ oligomers [16, 18], we performed western blotting on head extracts using the 6E10 pan-Aβ antibody to examine LDS/SDS-stable Aβ assemblies (supplementary Fig. 3b, c). The major LDS/SDS-stable Aβ species for all transgenic lines was monomeric (1-mer, short exposure time, supplementary Fig. 3b), however, longer exposure time revealed the presence of specific 6E10-positive bands of higher MWs, corresponding to the apparent size of Aβ dimers (2-mer, 8 kDa), trimers (3-mer, 12 kDa) and tetramers (4-mer, 16 kDa). Interestingly, the oligomeric profile was different for the investigated Aβ species, with a relative abundance of dimers for the Aβ42 line and of trimers for the Aβ43 line, while no LDS/SDS-stable low-MW Aβ oligomers could be observed in Aβ40-expressing flies (supplementary Fig. 3b, c).
Noteworthy, next to its propensity to self-aggregate, Aβ43 has also been implicated in the seeding of other Aβ peptides based on in vitro data and co-localisation studies [29, 38], an effect potentially enhancing overall Aβ toxicity. To test whether this process occurred in vivo, we used our Drosophila transgenic lines to stoichiometrically induce Aβ43 expression together with that of the non-toxic Aβ40 and to evaluate whether this combination would trigger toxicity. To that end, we titrated down Aβ43 levels by halving transgene copy number from two copies to one, leading to no detectable toxicity in terms of climbing ability (1×Aβ43, Fig. 6a) or median lifespan (1×Aβ43, Fig. 6b), similar to the effects obtained following halved expression of Aβ40 (1×Aβ40, Fig. 6a, b). While combining Aβ40 with Aβ40 itself (Aβ40+Aβ40) did not lead to any phenotypic changes as compared to Aβ40 alone (p > 0.05, Fig. 6a, b), we observed that the combination of low levels of both Aβ40 and Aβ43 induced synergistic toxic effects both on climbing ability (****p < 0.0001 vs. non-induced-Aβ40+Aβ43 controls and vs. all other induced lines at day 25, two-way ANOVA, Fig. 6a) and survival (median lifespan: −15.7 % of the non-induced controls, p < 0.0001, log-rank test, Fig. 6b), suggesting that Aβ43 was able to trigger toxicity from Aβ40 peptides in vivo, despite the ordinarily harmless nature of the latter. We used the same experimental setup to co-express Aβ40 with Aβ42 (supplementary Fig. 4), which led to drastic detrimental effects, both on climbing ability (****p < 0.0001 vs. non-induced-Aβ40+Aβ42 controls and vs. all other induced lines at day 18, two-way ANOVA, supplementary Fig. 4a) and survival (median lifespan: −46.7 % of the non-induced controls, p < 0.0001, log-rank test, supplementary Fig. 4b). Even though both Aβ40+Aβ42 and Aβ40+Aβ43 combinations led to significant synergistic toxic effects in the fly nervous system, it appeared that the former triggered stronger toxicity. We therefore investigated whether Aβ43 could modulate Aβ42 toxicity by comparing the effect of Aβ42+Aβ43 with that of Aβ42+Aβ42 on climbing ability and survival of flies (supplementary Fig. 5). We could observe a rather modest but yet significant reduction of toxicity in the combined Aβ42+Aβ43 line both in terms of climbing (**p < 0.01, induced-Aβ42+Aβ43 vs. induced-Aβ42+Aβ42, two-way ANOVA, supplementary Fig. 5a) and survival (p < 0.0001, induced-Aβ42+Aβ43 vs. induced-Aβ42+Aβ42, log-rank test, supplementary Fig. 5b). Importantly, we evaluated Aβ transcript levels among the lines in all three experiments and observed that they were comparable (p > 0.05, Aβ40+Aβ40 vs. Aβ40+Aβ43, Fig. 6c, Aβ40+Aβ40 vs. Aβ40+Aβ42, supplementary Fig. 4c, and Aβ42+Aβ42 vs. Aβ42+Aβ43, supplementary Fig. 5c, using Student’s t test), implying that the toxic interaction we observed between the Aβ isoforms was taking place at the protein level.
Aβ43 triggered toxicity from Aβ40. Climbing performance (a) and survival curves (b) of fly lines expressing 1×Aβ40 (grey), 1×Aβ43 (green) or the combination of Aβ40+Aβ40 (blue) or Aβ40+Aβ43 (navy blue) in adult neurons using the elavGS driver. The inset shows the percentage of median lifespan reduction vs. non-induced control. ****p < 0.0001 vs. non-induced controls, two-way ANOVA. c qRT-PCR analysis of Aβ mRNA levels from head extracts of Aβ40+Aβ40 and Aβ40+Aβ43 lines (p > 0.05, Student’s t test)
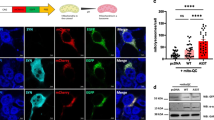
To verify this assumption and to further assess the potential seeding properties of Aβ43, we performed fractionation of Aβ according to its solubility in both Aβ40+Aβ43 and Aβ40+Aβ40 combined lines. Using the 6E10 pan-Aβ antibody (Fig. 7a), we observed a dramatic decrease of Aβ solubility in the combined Aβ40+Aβ43 condition as compared to the Aβ40+Aβ40 line, with insoluble Aβ species representing 57.89 ± 0.79 % of the total Aβ pool in the former line (****p < 0.0001, vs. Aβ40+Aβ40, Student’s t test, Fig. 7a, b). To determine whether this insoluble Aβ pool was holding Aβ40 species, which are intrinsically mainly soluble, we analysed the soluble and insoluble protein fractions using an antibody specifically targeting Aβ40 species (Fig. 8a). This revealed a progressive shift of Aβ40 species towards the insoluble fraction in the Aβ40+Aβ43 line as compared to the Aβ40+Aβ40 condition, the former having 33.37 ± 1.04 % of its Aβ40 species located in the SDS-insoluble fraction after 5 days of induction (vs. 8.64 ± 0.50 % for the Aβ40+Aβ40 line, ***p < 0.001, Student’s t test, Fig. 8b), while this proportion reached 87.11 ± 2.09 % after 14 days of induction (vs. 30.55 ± 2.49 % for the Aβ40+Aβ40 line, ****p < 0.0001, Student’s t test, Fig. 8c), implying that Aβ43 was a potent trigger of Aβ40 aggregation in vivo. To strengthen this observation, we performed immunofluorescence experiments on cryosections from Drosophila heads following Aβ over-expression in the compound eye, using an antibody specific to Aβ40. While the signal appeared predominantly diffuse in the Aβ40+Aβ40 expressing line, we observed a marked accumulation of Aβ40 species in the eye of the Aβ40+Aβ43 transgenic line (****p < 0.0001, Aβ40+Aβ40 vs. Aβ40+Aβ43, Student’s t test, Fig. 9), which is in line with the biochemical shift of Aβ40 species towards the insoluble protein fraction that we observed in the latter combination (Fig. 8). Altogether, our data indicate that Aβ43 species influence Aβ40 properties leading to its decreased solubility, which subsequently results in a significant enhancement of toxicity in vivo.
Combining Aβ40 with Aβ43 enhanced overall Aβ insolubility. a Fractionation and western blot of soluble (S) and insoluble (I) Aβ species retrieved from head extracts of Aβ40+Aβ40 and Aβ40+Aβ43 fly lines following induction in adult neurons (elavGS driver), using the pan-Aβ 6E10 antibody. b Quantification of the proportion of insoluble Aβ species extracted from Aβ40+Aβ40 and Aβ40+Aβ43 fly lines (****p < 0.0001, Student’s t test)
Combining Aβ40 with Aβ43 enhanced Aβ40 insolubility. a Western blot analysis of soluble (S) and insoluble (I) Aβ40 species retrieved from head extracts of Aβ40+Aβ40 and Aβ40+Aβ43 fly lines after 5 and 14 days of induction in adult neurons (elavGS driver), using an Aβ40-specific antibody. Quantification of the proportion of insoluble Aβ40 species after 5 (b) and 14 (c) days of induction (***p < 0.001 and ****p < 0.0001, Student’s t test)
Combining Aβ40 with Aβ43 enhanced Aβ40 immunoreactivity in fly compound eyes. Cryosections from heads of Aβ40+Aβ40- and Aβ40+Aβ43-expressing fly lines and of the GMR-Gal4 driver control line were probed with an Aβ40-specific antibody (green) and counterstained with DAPI (blue) and the number of Aβ40-positive events per eye section was quantified (****p < 0.0001, Student’s t test). Scale bar 50 µm
Discussion
While the amyloid load is built up by a mixture of Aβ peptides in AD brains [2], the respective involvement of these species in AD pathogenesis remains unsolved. Using inducible transgenic Drosophila lines expressing human Aβ, we demonstrated in the present study that Aβ43 peptides not only self-aggregate and lead to toxicity and neurodegeneration in vivo but also that they can trigger neurotoxicity from the otherwise innocuous Aβ40 peptides.
Our transgenic fly lines are based on the direct production of secretory Aβ species. Even though this model bypasses the secretase-based generation of Aβ from its APP precursor, potentially altering the normal trafficking route and sub-cellular localisation of APP and consequently of Aβ, the choice of directly expressing secretory forms of the different Aβ isoforms was crucial to ensure controlled and comparable levels of Aβ peptides produced upon induced expression in the fly nervous system. In addition, by generating transgenic lines using the attP/attB targeted integration system [20], we ensured that insertion of the different Aβ transgenes would take place into the same genomic locus, thereby reducing the risk of a differential promoter regulation due to a different genomic environment. In this way, we ensured that any observed toxic effect would not be a consequence of different transcript levels, which could have otherwise confounded the results, but would instead directly result from differential regulation occurring at the protein level. As an additional control to confirm the comparability of our transgenic lines, we measured the levels of Aβ peptide produced after a very short induction time, i.e. 4 h. We observed no significant difference in Aβ peptide levels between the Aβ transgenic fly lines at this stage, suggesting that translation from Aβ transcripts and therefore Aβ peptide synthesis was occurring at a similar rate, ruling out any potential differences in terms of translation efficiency among the analysed transgenic lines.
Interestingly, however, we observed that the amounts of total Aβ40, Aβ42 or Aβ43 retrieved from Drosophila heads became increasingly different with longer induction times. Since transcription and baseline Aβ peptide levels were comparable for all the lines, we hypothesized that such regulation at the peptide level was rather a consequence of altered clearance dynamics. Indeed, by means of a “switch-on/switch-off” experiment, where the RU486 inducer was removed after a period of 5 days, we could observe striking differences regarding the clearance of these Aβ isoforms. Such differential effects are likely to be linked to the unequal propensity of these Aβ species to form SDS-insoluble assemblies, as suggested by the differential kinetics of aggregation displayed by the Aβs. Indeed, among all three isoforms, we could observe that Aβ42 was the fastest isoform to generate SDS-insoluble species. Furthermore, even though the kinetics of Aβ43 aggregation in the fly nervous system was significantly slower than that of Aβ42 as observed after 1 day of induction, it was markedly faster than that of Aβ40, which mainly remained in a SDS-soluble state throughout the experiment. As compared to Aβ40, Aβ42 peptides hold two additional hydrophobic residues that greatly increase their propensity to aggregate [3, 15]. Moreover, recent in vitro studies [3, 15, 29] suggest that Aβ43, which bears an additional threonine in its C-terminal compared to Aβ42, displays even greater amyloidogenic properties than the latter. Our findings, however, suggest that when directly expressed in the fly nervous system, Aβ43 species shift towards an insoluble state with a slower kinetics than Aβ42. Although this would require further investigation, one might speculate that the molecular environment found in neuronal cells in vivo influences the kinetics of Aβ aggregation as compared to the behaviour of these peptides in vitro. It is important to note that, even when Aβ43 was found in a highly insoluble state in the fly nervous system, i.e. following 5 days of expression, the overall proportion of SDS-insoluble Aβ43 species was significantly lower than that of Aβ42, suggesting that the additional threonine in Aβ43 provides the peptide with an overall higher polarity and therefore lower amyloidogenicity than Aβ42. Altogether, our results suggest that Aβ40, Aβ42 and Aβ43 display a differential stability in vivo even when expressed at comparable levels, an effect that is likely related to their unequal propensity to form SDS-insoluble species, eventually influencing the ability of the cells to clear them.
Recent cell culture studies suggested that Aβ43 peptides are highly cytotoxic, significantly more so than Aβ40 peptides [1, 29]. In line with these data, we measured strong toxic effects resulting from Aβ43 expression in either Drosophila compound eyes or Drosophila adult neurons, as compared to the effects of Aβ40. However, we observed that in vivo, Aβ43 peptides were significantly less harmful than Aβ42 for all measured phenotypes, i.e. eye appearance, climbing ability, survival and neurodegeneration. Among the few cell culture studies that have so far investigated Aβ43, the relative cytotoxicity of the latter as compared to Aβ42 is still a matter of debate, some studies highlighting a higher toxicity of Aβ43 [23, 29], others suggesting that Aβ42 is more toxic [1]. Our in vivo results support the latter hypothesis, and it appears that in our system the observed differential toxic effects correlate with the unequal propensity of the Aβs to aggregate and therefore to be cleared from the cells, in agreement with several studies in Drosophila showing that Aβ toxicity is correlated with its propensity to form insoluble aggregates [11, 19, 34].
Nevertheless, given the increasing amount of data suggesting that soluble Aβ oligomers are important drivers of neurotoxicity in AD and AD models (for review, see [2]), together with recent data suggesting that Aβ43 assembles into soluble oligomers in vitro [31], we further analysed the potential occurrence of Aβ oligomers in our transgenic lines using the A11 oligomer-specific antibody. However, we could not highlight any PBS-soluble A11-positive oligomers in our lines, in agreement with previous studies analysing Aβ42 transgenic Drosophila [11]. However, since the A11 antibody does not detect low-MW Aβ oligomers, we used the pan-Aβ 6E10 antibody on western blot [18, 22] and could observe the presence of LDS/SDS-stable Aβ assemblies at the apparent MW of dimers, trimers and tetramers, the relative abundance of which differed among the investigated Aβ lines. While our results highlight the relative abundance of LDS/SDS-stable Aβ43 trimers and confirm published data that Aβ42 can form dimers in Drosophila [6, 11], further investigation will be required to decipher the potential contribution of these LDS/SDS-stable Aβ assemblies to the observed phenotypes.
Another important question in the context of AD, where several different Aβ species are found in brain deposits, is whether Aβ43 peptides could exacerbate neurotoxicity from other Aβ species in vivo. Therefore, we investigated whether Aβ43 could trigger toxicity from the non-toxic Aβ40 peptides in Drosophila. To that end, we reduced Aβ expression levels to a point where no toxicity could be detected, either on climbing ability or survival, by halving the Aβ transgene copy number from two to one. We combined one copy of each, Aβ40 with Aβ43 and as a control, Aβ40 with Aβ40 itself and, importantly, the measured overall Aβ transcript levels were comparable between these combinations. The Aβ40+Aβ43 combination was significantly more toxic on one hand than Aβ43 alone, despite identical Aβ43 transgene copy number, and on the other hand than the Aβ40+Aβ40 combination, even though the overall Aβ transcript levels did not differ between the conditions. At the protein level, the combination of Aβ40 with Aβ43 increased the proportion of SDS-insoluble Aβ40 species as compared to the Aβ40+Aβ40 line, and shifted the overall Aβ content towards a more insoluble state, suggesting that toxicity arose from an increased propensity to form insoluble structures in the Aβ40+Aβ43 combined line. Together with published observations that Aβ43 peptides can be found in plaque cores in AD brains [29, 36] and that they deposit earlier than other Aβ species in the brain of mouse models of AD [38], our data suggest that Aβ43 is involved in the titration of Aβ40 and potentially of other Aβ peptides into insoluble structures, therefore acting as a nucleating factor in AD brains. Our results, however, suggest that Aβ43 does not further exacerbate toxicity of the already extremely harmful Aβ42 in Drosophila.
In summary, our findings indicate a remarkable pathogenicity of Aβ43 peptides, not only because they self-aggregate and exert potent neurotoxic effects in vivo, but also because of their ability to trigger aggregation and toxicity from other Aβ species. Our results delineate the important contribution of Aβ43 peptides to the pathological events leading to neurodegeneration in AD, and suggest that these species represent a relevant target for the treatment of this disease.
References
Barucker C, Harmeier A, Weiske J, Fauler B, Albring KF, Prokop S et al (2014) Nuclear translocation uncovers the amyloid peptide Abeta42 as a regulator of gene transcription. J Biol Chem 289:20182–20191. doi:10.1074/jbc.M114.564690
Benilova I, Karran E, De Strooper B (2012) The toxic Abeta oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat Neurosci 15:349–357. doi:10.1038/nn.3028
Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB (2003) Amyloid beta-protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc Natl Acad Sci USA 100:330–335. doi:10.1073/pnas.222681699
Conicella AE, Fawzi NL (2014) The C-terminal threonine of Abeta43 nucleates toxic aggregation via structural and dynamical changes in monomers and protofibrils. Biochemistry 53:3095–3105. doi:10.1021/bi500131a
Cowan CM, Shepherd D, Mudher A (2010) Insights from Drosophila models of Alzheimer’s disease. Biochem Soc Trans 38:988–992. doi:10.1042/BST0380988
Crowther DC, Kinghorn KJ, Miranda E, Page R, Curry JA, Duthie FA et al (2005) Intraneuronal Abeta, non-amyloid aggregates and neurodegeneration in a Drosophila model of Alzheimer’s disease. Neuroscience 132:123–135. doi:10.1016/j.neuroscience.2004.12.025
Finelli A, Kelkar A, Song HJ, Yang H, Konsolaki M (2004) A model for studying Alzheimer’s Abeta42-induced toxicity in Drosophila melanogaster. Mol Cell Neurosci 26:365–375. doi:10.1016/j.mcn.2004.03.001
Franceschini N, Kirschfeld K, Minke B (1981) Fluorescence of photoreceptor cells observed in vivo. Science 213:1264–1267
Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ (2003) Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci USA 100:4078–4083. doi:10.1073/pnas.0737556100
Hao LY, Giasson BI, Bonini NM (2010) DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc Natl Acad Sci USA 107:9747–9752. doi:10.1073/pnas.0911175107
Iijima K, Chiang HC, Hearn SA, Hakker I, Gatt A, Shenton C et al (2008) Abeta42 mutants with different aggregation profiles induce distinct pathologies in Drosophila. PLoS ONE 3:e1703. doi:10.1371/journal.pone.0001703
Iijima K, Liu HP, Chiang AS, Hearn SA, Konsolaki M, Zhong Y (2004) Dissecting the pathological effects of human Abeta40 and Abeta42 in Drosophila: a potential model for Alzheimer’s disease. Proc Natl Acad Sci USA 101:6623–6628. doi:10.1073/pnas.0400895101
Iizuka T, Shoji M, Harigaya Y, Kawarabayashi T, Watanabe M, Kanai M et al (1995) Amyloid beta-protein ending at Thr43 is a minor component of some diffuse plaques in the Alzheimer’s disease brain, but is not found in cerebrovascular amyloid. Brain Res 702:275–278
Jackson GR, Salecker I, Dong X, Yao X, Arnheim N, Faber PW et al (1998) Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron 21:633–642
Jarrett JT, Berger EP, Lansbury PT Jr (1993) The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer’s disease. Biochemistry 32:4693–4697
Kayed R, Glabe CG (2006) Conformation-dependent anti-amyloid oligomer antibodies. Methods Enzymol 413:326–344. doi:10.1016/S0076-6879(06)13017-7
Keller L, Welander H, Chiang HH, Tjernberg LO, Nennesmo I, Wallin AK et al (2010) The PSEN1 I143T mutation in a Swedish family with Alzheimer’s disease: clinical report and quantification of Abeta in different brain regions. Eur J Hum Genet 18:1202–1208. doi:10.1038/ejhg.2010.107
Lesne SE, Sherman MA, Grant M, Kuskowski M, Schneider JA, Bennett DA et al (2013) Brain amyloid-beta oligomers in ageing and Alzheimer’s disease. Brain 136:1383–1398. doi:10.1093/brain/awt062
Luheshi LM, Tartaglia GG, Brorsson AC, Pawar AP, Watson IE, Chiti F et al (2007) Systematic in vivo analysis of the intrinsic determinants of amyloid Beta pathogenicity. PLoS Biol 5:e290. doi:10.1371/journal.pbio.0050290
Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N (2008) Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 40:476–483. doi:10.1038/ng.101
McLaurin J, Kierstead ME, Brown ME, Hawkes CA, Lambermon MH, Phinney AL et al (2006) Cyclohexanehexol inhibitors of Abeta aggregation prevent and reverse Alzheimer phenotype in a mouse model. Nat Med 12:801–808. doi:10.1038/nm1423
Meli G, Lecci A, Manca A, Krako N, Albertini V, Benussi L et al (2014) Conformational targeting of intracellular Abeta oligomers demonstrates their pathological oligomerization inside the endoplasmic reticulum. Nat Commun 5:3867. doi:10.1038/ncomms4867
Meng P, Yoshida H, Tanji K, Matsumiya T, Xing F, Hayakari R et al (2014) Carnosic acid attenuates apoptosis induced by amyloid-beta 1-42 or 1-43 in SH-SY5Y human neuroblastoma cells. Neurosci Res. doi:10.1016/j.neures.2014.12.003
Mizielinska S, Grönke S, Niccoli T, Ridler C, Clayton E, Devoy A et al (2014) C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345:1192–1194
Morais VA, Haddad D, Craessaerts K, De Bock PJ, Swerts J, Vilain S et al (2014) PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science 344:203–207. doi:10.1126/science.1249161
Osterwalder T, Yoon KS, White BH, Keshishian H (2001) A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA 98:12596–12601. doi:10.1073/pnas.221303298
Parvathy S, Davies P, Haroutunian V, Purohit DP, Davis KL, Mohs RC et al (2001) Correlation between Abetax-40-, Abetax-42-, and Abetax-43-containing amyloid plaques and cognitive decline. Arch Neurol 58:2025–2032
Rogers I, Kerr F, Martinez P, Hardy J, Lovestone S, Partridge L (2012) Ageing increases vulnerability to abeta42 toxicity in Drosophila. PLoS One 7:e40569. doi:10.1371/journal.pone.0040569
Saito T, Suemoto T, Brouwers N, Sleegers K, Funamoto S, Mihira N et al (2011) Potent amyloidogenicity and pathogenicity of Abeta43. Nat Neurosci 14:1023–1032. doi:10.1038/nn.2858
Sandebring A, Welander H, Winblad B, Graff C, Tjernberg LO (2013) The pathogenic abeta43 is enriched in familial and sporadic Alzheimer disease. PLoS One 8:e55847. doi:10.1371/journal.pone.0055847
Seither KM, McMahon HA, Singh N, Wang H, Cushman-Nick M, Montalvo GL et al (2014) Specific aromatic foldamers potently inhibit spontaneous and seeded Abeta42 and Abeta43 fibril assembly. Biochem J. doi:10.1042/BJ20131609
Selkoe DJ (2001) Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev 81:741–766
Sofola O, Kerr F, Rogers I, Killick R, Augustin H, Gandy C et al (2010) Inhibition of GSK-3 ameliorates Abeta pathology in an adult-onset Drosophila model of Alzheimer’s disease. PLoS Genet 6:e1001087. doi:10.1371/journal.pgen.1001087
Speretta E, Jahn TR, Tartaglia GG, Favrin G, Barros TP, Imarisio S et al (2012) Expression in Drosophila of tandem amyloid beta peptides provides insights into links between aggregation and neurotoxicity. J Biol Chem 287:20748–20754. doi:10.1074/jbc.M112.350124
Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL et al (2001) Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413:739–743. doi:10.1038/35099568
Welander H, Franberg J, Graff C, Sundstrom E, Winblad B, Tjernberg LO (2009) Abeta43 is more frequent than Abeta40 in amyloid plaque cores from Alzheimer disease brains. J Neurochem 110:697–706. doi:10.1111/j.1471-4159.2009.06170.x
Younkin SG (1998) The role of A beta 42 in Alzheimer’s disease. J Physiol Paris 92:289–292
Zou K, Liu J, Watanabe A, Hiraga S, Liu S, Tanabe C et al (2013) Abeta43 is the earliest-depositing Abeta species in APP transgenic mouse brain and is converted to Abeta41 by two active domains of ACE. Am J Pathol 182:2322–2331. doi:10.1016/j.ajpath.2013.01.053
Acknowledgments
We thank A. Bratic for helpful discussions and O. Hendrich for technical advice. This work was supported by the Max Planck Society, the Toxic Protein Conformations and Ageing Consortium of the Max Planck Society, and by the Wellcome Trust (WT098565). M. K. G. received support from the Cologne Graduate School of Ageing Research.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
401_2015_1419_MOESM1_ESM.pdf
Supplementary material 1: Fig. 1. Quantification of eye phenotypes. Quantification of the extent of eye roughening induced by the constitutive expression of Aβ40, Aβ42 or Aβ43 in the fly compound eye. Results are expressed as the percentage of analysed flies (moderate phenotype: *p < 0.05: Aβ43 vs. Aβ42; **p < 0.01: Aβ43 vs. Aβ40 and Aβ43 vs. GMR control; strong phenotype: #p < 0.05, Aβ43 vs. Aβ42; ##p < 0.01, Aβ42 vs. Aβ40 and Aβ42 vs. GMR control using Fisher’s exact test). (PDF 56 kb)
401_2015_1419_MOESM2_ESM.pdf
Supplementary material 2: Supplementary Fig. 2. Control experiments for the elavGS driver line. Climbing ability (a) and survival curves (b) of the elavGS driver control were analysed (RU486: plain lines, control food: dotted lines). The inset shows the percentage of median lifespan reduction vs. non-induced control. (PDF 65 kb)
401_2015_1419_MOESM3_ESM.pdf
Supplementary material 3: Supplementary Fig. 3. Characterization of Aβ oligomers. a. Dot blot analysis (left) of PBS-soluble fractions retrieved from head extracts of 5 days-old Aβ40, Aβ42 and Aβ43 transgenic flies and elavGS controls using the A11 oligomer-specific antibody. Quantification of A11 immunoreactivity is shown on the right panel. b. Heads of 14 days-old Aβ40, Aβ42 and Aβ43 transgenic flies and elavGS controls were directly extracted into LDS loading buffer and analysed by western blot using the pan-Aβ 6E10 antibody. Short exposure time (top panel) showed the levels of monomeric Aβ (1-mer) while longer exposure (bottom panel) revealed higher Aβ bands at the apparent size of dimers (2-mer), trimers (3-mer) and tetramers (4-mer) and pointed to a differential oligomeric profile between the transgenic lines. No oligomeric Aβ species could be observed in extracts from the Aβ40-expressing flies and no signal was detected in the negative control in this range of molecular weight (elavGS driver line). Western blot for Actin reveals a comparable loading for all the lanes. c. The densitometry profiles show the relative intensity of the Aβ bands for Aβ42 and Aβ43 transgenics. (PDF 6345 kb)
401_2015_1419_MOESM4_ESM.pdf
Supplementary material 4: Supplementary Fig. 4. Aβ 42 triggered toxicity from Aβ 40 . a and b. Climbing performance (a) and survival curves (b) of fly lines expressing 1xAβ40 (grey), 1xAβ42 (violet) or the combination of Aβ40+Aβ40 (blue) or Aβ40+Aβ42 (orange) in adult neurons using the elavGS driver. The inset shows the percentage of median lifespan reduction vs. non-induced control. ****p < 0.0001 vs. non-induced controls, two-way ANOVA. c. qRT-PCR analysis of Aβ mRNA levels from head extracts of Aβ40+Aβ40 and Aβ40+Aβ42 lines (p > 0.05, Student’s t test). (PDF 141 kb)
401_2015_1419_MOESM5_ESM.pdf
Supplementary material 5: Supplementary Fig. 5. Aβ 43 did not exacerbate Aβ 42 -induced toxicity. a and b. Climbing performance (a) and survival curves (b) of fly lines expressing the combination of either Aβ42+Aβ42 (purple) or Aβ42+Aβ43 (pink) in adult neurons using the elavGS driver. The inset shows the percentage of median lifespan reduction vs. non-induced control. **p < 0.01, induced-Aβ42+Aβ42 vs. induced-Aβ42+Aβ43 at day 11, two-way ANOVA. c. qRT-PCR analysis of Aβ mRNA levels from head extracts of Aβ42+Aβ42 and Aβ42+Aβ43 lines (p > 0.05, Student’s t test). (PDF 133 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Burnouf, S., Gorsky, M.K., Dols, J. et al. Aβ43 is neurotoxic and primes aggregation of Aβ40 in vivo. Acta Neuropathol 130, 35–47 (2015). https://doi.org/10.1007/s00401-015-1419-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-015-1419-y