Abstract
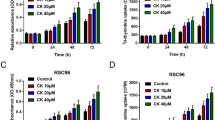
Schwann cells (SCs) proliferation is crucial for nerve regeneration following nerve injury. This study aims to investigate effects of interleukin-22 (IL-22) on SCs proliferation in vitro, as well as the corresponding mechanism. Rat SCs were treated with 100 ng/ml rat IL-22 for 48 h, and cell proliferation and apoptosis were detected using fluorescent staining and flow cytometry. After transcriptome sequencing, raw reads were filtered and mapped to reference genome rn5. Then, differentially expressed genes (DEGs) and long non-coding RNAs (DElncRNAs) between IL-22 and control groups were identified (tool: Cuffdiff). Functional and pathway enrichment analyses were performed (tool: GOFunction), and protein–protein interaction (PPI) network was constructed (tool: STRING and Cytoscape). Furthermore, Pearson’s correlations between DEGs and DElncRNAs were analyzed, and regulatory network of DEGs, DElncRNAs, and transcription factors (TFs) was constructed. IL-22 significantly inhibited proliferation (p value < 0.05) and promoted apoptosis of Schwann cells. Totally, 932 DEGs and 118 DElncRNAs were identified, among which Ccl2 and Ccna2 were hub genes in PPI network. Up-regulated DEGs were enriched in apoptosis related terms, whereas down-regulated DEGs were enriched in proliferation related terms. DElncRNAs like NONRATT023505, NONRATG020400, and NONRATT022748 were correlated with multiple DEGs enriched in cell cycle and division. Moreover, up-regulated TFs Egr1, Cebpd, and Atf4 play crucial roles in regulatory network, and NONRATG020400-Cebpd-Ccl2, NONRATT023505/NONRATT022748-Atf4-Ccna2, and NONRATT022748-Egr1-Id1/Aldoc/Eno2/F3/Serpine1 regulatory pathways were identified in SCs after IL-22 treatment. IL-22 might influence SCs proliferation and apoptosis via regulating lncRNA–TF–gene pathways in SCs. However, more studies are required to confirm these results.




Similar content being viewed by others
References
Arthur-Farraj PJ, Latouche M, Wilton DK, Quintes S, Chabrol E, Banerjee A, Woodhoo A, Jenkins B, Rahman M, Turmaine M, Wicher GK, Mitter R, Greensmith L, Behrens A, Raivich G, Mirsky R, Jessen KR (2012) c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron 75(4):633–647. doi:10.1016/j.neuron.2012.06.021
Kanno H, Pressman Y, Moody A, Berg R, Muir EM, Rogers JH, Ozawa H, Itoi E, Pearse DD, Bunge MB (2014) Combination of engineered Schwann cell grafts to secrete neurotrophin and chondroitinase promotes axonal regeneration and locomotion after spinal cord injury. J Neurosci 34(5):1838–1855. doi:10.1523/jneurosci.2661-13.2014
Ghosh M, Tuesta LM, Puentes R, Patel S, Melendez K, El Maarouf A, Rutishauser U, Pearse DD (2012) Extensive cell migration, axon regeneration, and improved function with polysialic acid-modified Schwann cells after spinal cord injury. Glia 60(6):979–992. doi:10.1002/glia.22330
Susuki K (2014) Schwann Cell-Dependent Regulation of Peripheral Nerve Injury and Repair. In: Schwann Cell Development and Pathology. Springer., pp 69–79
Kobayashi M, Ishibashi S, Tomimitsu H, Yokota T, Mizusawa H (2012) Proliferating immature Schwann cells contribute to nerve regeneration after ischemic peripheral nerve injury. J Neuropathol Exp Neurol 71(6):511–519. doi:10.1097/NEN.0b013e318257fe7b
Chang HM, Liu CH, Hsu WM, Chen LY, Wang HP, Wu TH, Chen KY, Ho WH, Liao WC (2014) Proliferative effects of melatonin on Schwann cells: implication for nerve regeneration following peripheral nerve injury. J Pineal Res 56(3):322–332. doi:10.1111/jpi.12125
Tang X, Wang Y, Zhou S, Qian T, Gu X (2013) Signaling pathways regulating dose-dependent dual effects of TNF-alpha on primary cultured Schwann cells. Mol Cell Biochem 378(1–2):237–246. doi:10.1007/s11010-013-1614-x
Temporin K, Tanaka H, Kuroda Y, Okada K, Yachi K, Moritomo H, Murase T, Yoshikawa H (2008) IL-1beta promotes neurite outgrowth by deactivating RhoA via p38 MAPK pathway. Biochem Biophys Res Commun 365(2):375–380. doi:10.1016/j.bbrc.2007.10.198
Conti G, De Pol A, Scarpini E, Vaccina F, De Riz M, Baron P, Tiriticco M, Scarlato G (2002) Interleukin-1 beta and interferon-gamma induce proliferation and apoptosis in cultured Schwann cells. J Neuroimmunol 124(1–2):29–35
Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA (2006) Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med 203(10):2271–2279. doi:10.1084/jem.20061308
Li S, Yu M, Li H, Zhang H, Jiang Y (2012) IL-17 and IL-22 in cerebrospinal fluid and plasma are elevated in Guillain-Barre syndrome. Mediators Inflamm 2012:260473. doi:10.1155/2012/260473
Li S, Jin T, Zhang HL, Yu H, Meng F, Concha Quezada H, Zhu J (2014) Circulating Th17, Th22, and Th1 cells are elevated in the Guillain-Barre syndrome and downregulated by IVIg treatments. Mediators Inflamm 2014:740947. doi:10.1155/2014/740947
Holoch D, Moazed D (2015) RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16(2):71–84. doi:10.1038/nrg3863
Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y (2014) Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep 31(1):358–364. doi:10.3892/or.2013.2850
Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY (2013) Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 13:461. doi:10.1186/1471-2407-13-461
Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY (2013) A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2, e00762. doi:10.7554/eLife.00762
Doddrell RD, Dun XP, Moate RM, Jessen KR, Mirsky R, Parkinson DB (2012) Regulation of Schwann cell differentiation and proliferation by the Pax-3 transcription factor. Glia 60(9):1269–1278. doi:10.1002/glia.22346
Li A, Gan Y, Wang R, Liu Y, Ma T, Huang M, Cui X (2014) IL-22 up-regulates beta-defensin-2 expression in human alveolar epithelium via STAT3 but not NF-kappaB signaling pathway. Inflammation. doi:10.1007/s10753-014-0083-z
Qu D, Wang G, Wang Z, Zhou L, Chi W, Cong S, Ren X, Liang P, Zhang B (2011) 5-Ethynyl-2′-deoxycytidine as a new agent for DNA labeling: detection of proliferating cells. Anal Biochem 417(1):112–121. doi:10.1016/j.ab.2011.05.037
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7(2), e30619. doi:10.1371/journal.pone.0030619
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14(4):R36
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7(3):562–578
Liu C, Bai B, Skogerbø G, Cai L, Deng W, Zhang Y, Bu D, Zhao Y, Chen R (2005) NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res 33(suppl 1):D112–D115
Raivo K (2012) pheatmap: Pretty Heatmaps. R package version 0.7. 4
Wang J, Zhou X, Zhu J, Gu Y, Zhao W, Zou J, Guo Z (2012) GO-function: deriving biologically relevant functions from statistically significant functions. Brief Bioinform 13(2):216–227. doi:10.1093/bib/bbr041
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, Von Mering C, Jensen LJ (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41(Database issue):D808–D815. doi:10.1093/nar/gks1094
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27(3):431–432. doi:10.1093/bioinformatics/btq675
Sedgwick P (2012) Pearson’s correlation coefficient. Br Med J 345
Jiang C, Xuan Z, Zhao F, Zhang MQ (2007) TRED: a transcriptional regulatory element database, new entries and other development. Nucleic Acids Res 35(suppl 1):D137–D140
Fury W, Batliwalla F, Gregersen PK, Li W (2006) Overlapping probabilities of top ranking gene lists, hypergeometric distribution, and stringency of gene selection criterion. Conf Proc IEEE Eng Med Biol Soc 1:5531–5534. doi:10.1109/iembs.2006.260828
Hinojosa AE, Garcia-Bueno B, Leza JC, Madrigal JL (2011) CCL2/MCP-1 modulation of microglial activation and proliferation. J Neuroinflammation 8:77. doi:10.1186/1742-2094-8-77
Chen TA, Wang JL, Hung SW, Chu CL, Cheng YC, Liang SM (2011) Recombinant VP1, an Akt inhibitor, suppresses progression of hepatocellular carcinoma by inducing apoptosis and modulation of CCL2 production. PLoS One 6(8), e23317. doi:10.1371/journal.pone.0023317
Nerlov C (2007) The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol 17(7):318–324. doi:10.1016/j.tcb.2007.07.004
Liu G, Su L, Hao X, Zhong N, Zhong D, Singhal S, Liu X (2012) Salermide up-regulates death receptor 5 expression through the ATF4-ATF3-CHOP axis and leads to apoptosis in human cancer cells. J Cell Mol Med 16(7):1618–1628. doi:10.1111/j.1582-4934.2011.01401.x
Galehdar Z, Swan P, Fuerth B, Callaghan SM, Park DS, Cregan SP (2010) Neuronal apoptosis induced by endoplasmic reticulum stress is regulated by ATF4-CHOP-mediated induction of the Bcl-2 homology 3-only member PUMA. J Neurosci 30(50):16938–16948. doi:10.1523/jneurosci.1598-10.2010
Stewart HJ, Zoidl G, Rossner M, Brennan A, Zoidl C, Nave KA, Mirsky R, Jessen KR (1997) Helix-loop-helix proteins in Schwann cells: a study of regulation and subcellular localization of Ids, REB, and E12/47 during embryonic and postnatal development. J Neurosci Res 50(5):684–701
Diaz-Ramos A, Roig-Borrellas A, Garcia-Melero A, Lopez-Alemany R (2012) alpha-Enolase, a multifunctional protein: its role on pathophysiological situations. J Biomed Biotechnol 2012:156795. doi:10.1155/2012/156795
Xie B, Wang C, Zheng Z, Song B, Ma C, Thiel G, Li M (2011) Egr-1 transactivates Bim gene expression to promote neuronal apoptosis. J Neurosci 31(13):5032–5044. doi:10.1523/jneurosci.5504-10.2011
Zins K, Pomyje J, Hofer E, Abraham D, Lucas T, Aharinejad S (2014) Egr-1 upregulates Siva-1 expression and induces cardiac fibroblast apoptosis. Int J Mol Sci 15(1):1538–1553. doi:10.3390/ijms15011538
Wang C, Husain K, Zhang A, Centeno BA, Chen DT, Tong Z, Sebti SM, Malafa MP (2015) EGR-1/Bax pathway plays a role in vitamin E delta-tocotrienol-induced apoptosis in pancreatic cancer cells. J Nutr Biochem 26(8):797–807. doi:10.1016/j.jnutbio.2015.02.008
Han MH, Kim GY, Yoo YH, Choi YH (2013) Sanguinarine induces apoptosis in human colorectal cancer HCT-116 cells through ROS-mediated Egr-1 activation and mitochondrial dysfunction. Toxicol Lett 220(2):157–166. doi:10.1016/j.toxlet.2013.04.020
Acknowledgments
This study was supported by National Natural Science Foundations of China (81200924, 81200953 and 81400152); The Natural Science Foundation of Shanghai Municipality, China (12ZR1427400) and The Youth Fund of the Shanghai Health Bureau, China (Mingyuan Liu 2011).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Shengming Xu and Junping Ao are regarded as co-first author.
Rights and permissions
About this article
Cite this article
Xu, S., Ao, J., Gu, H. et al. IL-22 Impedes the Proliferation of Schwann cells: Transcriptome Sequencing and Bioinformatics Analysis. Mol Neurobiol 54, 2395–2405 (2017). https://doi.org/10.1007/s12035-016-9699-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-9699-3