Abstract
The extracellular signal-regulated protein kinase 5 (ERK5) is a mitogen-activated protein kinase that phosphorylates and regulates various transcription factors in response to growth factors and extracellular stresses. To address its biological function during the development of the peripheral nervous system (PNS), we have engineered a novel model of sympathetic neurons in which the erk5 gene can be deleted in vitro. Our data provide for the first time genetic evidence that ERK5 is required to mediate the survival response of neurons to nerve growth factor. Increased cell death associated with the loss of ERK5 is caused by elevated expression of the BH3-only members of the Bcl-2 family, Bad and Bim. Further investigation indicated that ERK5 suppresses the transcription of the bad and the bim genes by Ca2+/cAMP response element-binding protein and Forkhead box O3a, respectively. Consistently, we found that the phosphorylation of both p90 ribosomal S6 kinase and protein kinase B is impaired in neurons lacking ERK5. Together these findings reveal a novel signaling mechanism that promotes neuronal survival during the development of the PNS.
Similar content being viewed by others
Main
During the development of the brain approximately half of the neurons formed by neurogenesis die by apoptosis before adulthood because of limiting amounts of trophic factors.1 The molecular mechanism underlying this neuronal loss has been greatly facilitated by the phenotypic analysis of nerve growth factor (NGF)-dependent sympathetic neurons lacking specific members of the Bcl-2 family.2 For example, whereas Bax is essential for NGF withdrawal-induced apoptosis,3 there is functional redundancy between the BH3-only proteins Bad and Bid.4 Unlike bad−/− neurons and bid−/− neurons that do not exhibit any abnormal phenotype, the targeted deletion of bim partially protects sympathetic neurons against trophic factor withdrawal.5 Further studies have shown that increased expression of Bim by the transcription factors c-Jun and Forkhead box O3a (Foxo3a) is critical to trigger neuronal apoptosis.6, 7
Similarly, de novo protein synthesis is a prerequisite for trophic factor-induced survival. This is exemplified in vivo by the decreased number of sensory and sympathetic neurons in the brain of Ca2+/cAMP response element-binding protein (CREB)−/− mice.8 CREB is a prosurvival transcription factor that acts as both an activator and a repressor of gene expression in the nervous system.9 The transcriptional activity of CREB is increased upon phosphorylation at serine 133 by numerous protein kinases, including p90 ribosomal S6 kinase (RSK), a downstream target of extracellular-regulated protein kinases 1 and 2 (ERK1/2) and ERK5.10, 11 The specific requirement of ERK5, but not of ERK1/2, to activate CREB in distally stimulated NGF-dependent sensory neurons provided the first evidence that ERK5 was essential for promoting trophic factor-induced neuronal survival.12 The role of ERK5 in mediating the survival of neurons in the central nervous system by the activation of the myocyte enhancer factor 2 transcription factor has since been reported.13, 14
ERK5 is a nonredundant mitogen-activated protein kinase (MAPK) stimulated in response to growth factors and cellular stresses by the MAPK/ERK kinase 5 (MEK5).15 As its cloning in 1995, the lack of biological tools, including specific inhibitors, have made it one of the least studied MAPK subfamilies. Consequently, little is known about the downstream targets of ERK5 and therefore the biochemical mechanisms that mediate the effect of ERK5 remain largely unidentified. To advance the current knowledge, we have developed a novel model of primary cultures of sympathetic neurons in which the erk5 gene can be deleted in vitro. Here we provide genetic evidence that ERK5 is required for neuronal survival by suppressing Bad and Bim expression by CREB and Foxo3a, respectively.
Results
Deletion of the erk5 gene causes apoptosis
The role of ERK5 in NGF-mediated neuronal survival was examined by testing the effect of erk5 gene deletion in sympathetic neurons. Homozygous erk5loxP superior cervical ganglion (SCG) neurons were infected with an adenovirus encoding Cre recombinase (Cre) or the green fluorescence protein (GFP; Figure 1a). Immunofluorescence staining confirmed that adenoviruses at 100 multiplicity of infection (MOI)-infected neurons with 100% efficiency (Figure 1a). Genomic PCR analysis using specific primers flanking exon 3 revealed that infection with the Cre virus at 100 MOI for 24 h induced efficient recombination of the erk5 gene (Figure 1b). This correlated with the complete loss of the ERK5 protein after 48 h, as observed by immunoblot analysis of the cell lysates using a specific antibody to ERK5 (Figure 1b). The retarded migration of ERK5 following SDS-polyacrylamide gel electrophoresis (PAGE) analysis of wild-type extracts was absent when the cells were incubated with the alkaline phosphatase CIP, suggesting that the upper band detected by immunoblot corresponded to a phosphorylated form of ERK5 (Figure 1c). Similarly, the electrophoretic mobility shift was abolished in SCG neurons cultured in the absence of NGF for 15 and 30 min (Figure 1c). The phosphorylation of ERK5 was restored 30 min after the readdition of NGF (Figure 1c). Together these results demonstrate that ERK5 is phosphorylated in SCG neurons incubated with NGF.
erk5 gene deletion sensitizes neurons to apoptosis. Homozygous flox (a–g) or wild-type (h) SCG neurons were not infected or infected with a control adenovirus (GFP) or with an adenovirus encoding Cre at 100 MOI. The cells were cultured for a further 36 h (a) or 48 h (b, d–h) in presence of NGF (50 ng/ml), unless indicated otherwise. (a) Immunofluorescence analysis of SCG neurons to detect GFP (green) and Cre (anti-Cre antibody, red) expression demonstrates that 100% of the cells were infected by the recombinant adenoviruses. DNA was stained with DAPI (blue). Scale bar, 25 μM. (b, i) Genomic DNA isolated from the cells was amplified by PCR with primers specific for the erk5 gene. erk5f and erk5 correspond to the erk5-flox and disrupted allele, respectively. (ii) Proteins were extracted and analyzed by immunoblot using specific antibodies to ERK5 and to tubulin. (c) Extracts were analyzed for ERK5 expression by immunoblot. The detection of tubulin expression was performed to monitor protein loading. Where indicated, NGF was removed (− NGF) for 15 and 30 min and readded (+ NGF) for 30 min, before the cells being harvested. CIP treatment of the extract before analysis is indicated. Similar results were obtained in two independent experiments. (d) Extracts were analyzed for JNK expression and phosphorylation (P) by immunoblot. Where indicated, NGF was removed (−NGF) for 6 h before the cells being harvested. Similar results were obtained in two independent experiments. (e) Phase-contrast photomicrographs of representative fields of SCG neurons expressing GFP or Cre are shown. Scale bar, 25 μM. (f) SCG neurons were incubated with Hoechst 33342 and propidium iodide to distinguish viable, necrotic and apoptotic cells. Only neurons that had clearly segmented and condensed chromatin were counted as apoptotic. The classification criteria are shown in supplementary Figure 1. (g, h) Caspase-3 activity was measured by caspase assay. In some experiments, NGF was removed (0) for 18 h before the cells being harvested (f, g). The data, expressed as the mean±standard error (S.E.), were generated from three independent experiments performed in duplicate (f–h). *P⩽0.001 indicates a significant difference between GFP- and Cre-infected neurons. The electrophoretic mobility shift caused by the phosphorylation of ERK5 is indicated by an arrow
SCG neurons are dependent on trophic support for their survival. This is demonstrated by NGF withdrawal-induced phosphorylation of the proapoptotic c-Jun N-terminal protein kinase (JNK) MAPK (Figure 1d), as well as an increased number of nuclei displaying segmented and condensed chromatin (Supplementary Figure 1a and b). In addition, caspase-3 activity was elevated with a maximum at 24 h after NGF deprivation (Supplementary Figure 1c). Similarly, the absence of ERK5 for 48 h promoted morphological changes in cell shape (Figure 1e) and in chromatin structure (Figure 1f) typical of apoptotic cells, and significantly increased caspase-3 activity (Figure 1g). However, in contrast to NGF withdrawal, the loss of ERK5 did not increase JNK phophorylation (Figure 1d). The level of apoptotic death associated with ablation of ERK5 in the presence of NGF was comparable to that observed with the removal of NGF for 18 h (Figure 1f and g). Control experiments demonstrated that infection of wild-type SCG neurons with the Cre virus was not toxic to the cells (Figure 1h). Together these results indicate that ERK5 is a crucial mediator of the NGF prosurvival signal.
ERK1/2 and protein kinase B (PKB, also known as Akt) have previously been implicated in protecting neurons against stress.16 To establish the relative importance of ERK5, ERK1/2 and PKB in mediating NGF-dependent neuronal survival, we compared the effect of ERK5 deletion with the specific inhibition of ERK1/2 and PKB signaling. Incubation of the cells with either UO126 or wortmannin completely abolished the phosphorylation of ERK1/2 and of PKB at Thr308, but not that of ERK5, demonstrating the specificity of the drugs (Figure 2a). Conversely, ablation of ERK5 by infecting erk5loxP SCG neurons with Cre virus did not affect the phosphorylation of ERK1/2 or PKB at Thr308 (Figure 2a). Under conditions of adequate trophic support, loss of ERK5 or inhibition of ERK1/2 or PKB signaling, increased caspase-3 activity to a similar extent (P>0.05; Figure 2b). In contrast to our data, several studies have demonstrated that MEK inhibition has minimal effects on NGF-dependent neuronal survival.17, 18, 19, 20 Therefore, to confirm our results, we tested the effect of PD0325901, a novel noncompetitive inhibitor of MEK1 with greater potency than UO126.21 Like UO126, we found that PD0325901 specifically inhibited ERK1/2 phosphorylation (Figure 2c) and decreased the survival of SCG neurons incubated with NGF (Figure 2d). Consistent with the caspase-3 assay (Figure 2b) there was no significant difference in cell death exhibited by SCG neurons infected with Cre or incubated with PD0325901 or with wortmannin (Figure 2d). The effect of functional inhibition of ERK5 and ERK1/2, ERK5 and PKB, or ERK5, ERK1/2 and PKB signaling was additive (Figure 2b and d). Together, these results indicate that the ERK5, ERK1/2 and PKB pathways contribute similarly to the survival of NGF-dependent SCG neurons and that neither of them can fully substitute for the loss of the others.
ERK5, ERK1/2 and PKB are required to support the survival of NGF-dependent SCG neurons. Homozygous flox SCG neurons were infected with an adenovirus encoding GFP or Cre. The cells were cultured for a further 48 h in presence of NGF (50 ng/ml). Where indicated, the cells were treated with UO126 (10 μM), wortmannin (50 nM) or PD0325901 (25 nM) 6 h after the infection. The drugs were replaced every 12 h for the remaining time of the infection. (a, c) Extracts were analyzed for ERK5, ERK1/2 and PKB expression, and for phosphorylation (P) of ERK1/2 and of PKB at Thr308 by immunoblot. The electrophoretic mobility shift caused by the phosphorylation of ERK5 is indicated by an arrow. Similar results were obtained in two independent experiments. (b) Caspase-3 activity was measured by caspase assay. The data correspond to the mean±S.E. of three independent experiments performed in duplicate. (d) Cell survival was measured by LDH assay. The data correspond to the mean±range of two independent experiments performed in duplicate. *P<0.05 indicates a significant difference between GFP- and Cre-infected neurons or between Cre-infected neurons treated or not with the inhibitors. NS, indicates no significant difference (P>0.05)
ERK5 is required to inhibit Bad and Bim expression
To elucidate the mechanism by which the loss of ERK5 causes neuronal apoptosis we investigated the regulation of Bad, whose functional inhibition by ERK5 prevents apoptosis in endothelial cells.22 Immunoblot analysis using a Bad-specific antibody revealed that the expression of Bad was increased by around threefold after ablation of ERK5 (Figure 3a). Immunofluorescence studies confirmed that Bad was expressed at a much lower level in control neurons than in neurons lacking ERK5. Indeed, the exposure time used to generate the images was insufficient to detect Bad in LacZ-infected cells (Figure 3b). The staining of Bad displayed by Cre-infected erk5loxP neurons partially colocalized with the mitochondrial heat shock protein 70 (mHsp70; Figure 3b). This is consistent with the proapoptotic function of Bad at the mitochondria where it inhibits the activity of antiapoptotic Bcl-2 family members.23 Similar increases in the expression of the extra long and long Bim isoforms were detected following the loss of ERK5 (Figure 3c). In contrast, no marked difference was observed in the levels of Bid expression between GFP- and Cre-expressing erk5loxP SCG neurons up until 72 h postinfection (Figure 3d and data not shown). Furthermore, the loss of ERK5 did not cause Bid cleavage (data not shown).
ERK5 is required to suppress the expression of Bad and Bim. Homozygous flox SCG neurons were infected with a control adenovirus (GFP or LacZ) or with an adenovirus encoding Cre. In (e) and (f), the cells were infected 1 h later with lentiviruses encoding control, bim or bad shRNA. The neurons were cultured for a further 48 h in presence of NGF (50 ng/ml). (a, c–e) Extracts were analyzed for Bad, Bim and Bid expression by immunoblot. The detection of tubulin expression was performed to monitor protein loading. Images of Bad and Bim are from the same samples. Immunoblot signals were quantified with the ImageQuantifier software (BioImage, Jackson, MI, USA). The data correspond to the mean±S.E. of three independent experiments. *P<0.001 indicates a significant difference between GFP and Cre-infected neurons; NS, indicates no significant difference (P>0.05). (b) Immunofluorescence was performed with specific antibodies to Bad and mitochondrial mHsp70. Immune complexes were detected with secondary antibodies conjugated to Texas red (Bad) or fluorescein (mHsp70). DNA was stained with DAPI (blue). Scale bar, 5 μM. (f) Caspase-3 activity was measured by caspase assay. The data correspond to the mean±range of two independent experiments performed in duplicate
To determine the physiological significance of the increase in bad and bim expression we tested the effect of downregulating their expression in erk5−/− SCG neurons (Figure 3e and f). Neurons were infected with adenoviruses encoding GFP or Cre 1 h before incubating the cells with recombinant shRNA lentivirus targeting the murine bad or bim genes. A recombinant lentivirus encoding unspecific (ctrl) shRNA was used as a control to monitor the effect of lentivirus infection. Immunoblot analysis indicated that Bad and Bim shRNA efficiently and specifically prevented the increase in Bad and Bim expression caused by erk5 gene deletion (Figure 3e). The downregulation of either of these BH3-only proteins prevented the neuronal death associated with the loss of ERK5 (Figure 3f). Together these results demonstrate that Bad and Bim are required to induce the death of neurons lacking ERK5.
ERK5 controls the transcription of Bad and Bim by CREB and Foxo3a
To determine how ERK5 controls Bad and Bim expression, the level of their transcripts in SCG neurons expressing GFP or Cre was measured by quantitative real-time (RT)-PCR (Figure 4a). The upregulation of bad mRNA was transient and maximal 18 h after infection with Cre virus, a time that corresponded to a 70% reduction of the erk5 transcript. Expression of Cre for 36 h was necessary to detect increased bim mRNA. Control experiments demonstrated that the transcription of erk5, bad and bim genes was not affected in wild-type SCG neurons infected with the Cre virus (data not shown), strengthening our conclusion that ERK5 is required to downregulate the expression of Bad and Bim under adequate trophic factors conditions.
ERK5 regulates Bad and Bim transcription. (a) Homozygous flox SCG neurons were infected with an adenovirus encoding GFP or Cre. The cells were cultured in presence of NGF (50 ng/ml) for the indicated times. Total RNA was extracted and the amounts of erk5, bad and bim transcripts were measured by RT-PCR. (b, c) Homozygous flox SCG neurons were transiently transfected with a Bad or a Bim reporter luciferase plasmid and a pRL-Tk plasmid 20 or 2 h before being infected with an adenovirus encoding GFP or Cre for 18 and 36 h, respectively. The transcriptional activity was measured by the Dual-Luciferase reporter assay system. (d–f) PC6.3 cells were transiently cotransfected with a Bad or a Bim reporter luciferase plasmid and a pRL-Tk plasmid together with (+) or without (−) an expression vector encoding flag-tagged DN-ERK5. The following day the cells were cultured in differentiating medium without or with UO126 (10 μM) or wortmanin (50 nM), for a further 36 h. The inhibitors were replaced every 12 h. Cell extracts were analyzed for the expression of DN-ERK5, Bad and Bim, and for the phosphorylation (P) of ERK5 by immunoblot (d). The detection of tubulin expression was performed to monitor protein loading. The transcriptional activity was measured by the Dual-Luciferase reporter assay system (e, f). All the data correspond to the mean±S.E. of three independent experiments performed in duplicate
The transcriptional regulation of Bad and Bim by ERK5 was further examined by luciferase assay using reporter plasmids containing a fragment of the Bad (Bad-Luc) or of the Bim (Bim-Luc) promoter (Figure 4b and c). Increased luciferase activity in SCG neurons following the loss of ERK5 confirmed the negative effect of ERK5 on the bad and bim promoters (Figure 4b and c). Consistent with the quantitative RT-PCR time–course analysis (Figure 4a), Bad-luc was activated in neurons 18 h after infection, whereas 36 h was necessary to detect upregulation of Bim-luc activity.
Increased activity of Bad-luc and Bim-luc was also observed in differentiated PC6.3 overexpressing a dominant-negative (DN) form of ERK5 (Figure 4e and f). Immunoblot analysis using a phosphospecific antibody against ERK5 confirmed that ectopic expression of DN-ERK5 blocked the ability of MEK5 to activate ERK5 (Figure 4d). This correlated with increased Bad and Bim expression. The ability of UO126 and wortmannin to activate Bim-luc (Figure 4f), but not Bad-luc (Figure 4e), demonstrated that ERK1/2 and PKB contribute to downregulating the expression of Bim, but not of Bad, under adequate trophic factor conditions.
A number of response elements have been identified in the promoters of both the bad and the bim genes, including putative-binding sites for the transcription factors CREB and Foxo3a, which have both been reported to be downstream targets of the ERK5 signaling pathway.12, 24 Bad(mt)-luc and Bim(dm)-luc reporter constructs carrying deletions of the putative CREB and Foxo3a-binding sites, respectively, were not responsive to the loss of ERK5 (Figure 4b and c). Together, these results indicate that ERK5 suppresses Bad and Bim expression by its ability to stimulate CREB and to inhibit Foxo3a activity, respectively.
ERK5 regulates CREB and Foxo3a activity by RSK and PKB
Next, we found that SCG neurons lacking ERK5 (Figure 5a) or differentiated PC6.3 overexpressing DN-ERK5 (Figure 5b) exhibited impaired phosphorylation of RSK and CREB. Decreased phosphorylation of CREB in SCG neurons was transient with a maximum at 18 h after erk5 gene deletion. This indicates that CREB can be phosphorylated in the absence of ERK5 independently of RSK. This compensatory signaling mechanism could explain the transient upregulation of the bad transcript in SCG neurons following the loss of ERK5 (Figure 4a).
ERK5 regulates the transcription of bad by RSK-dependent CREB phosphorylation. (a) Homozygous flox SCG neurons were infected with an adenovirus encoding GFP or Cre. The cells were cultured in presence of NGF (50 ng/ml) for the indicated times. (b–d) PC6.3 cells were transiently transfected with a wild-type or with a CRE-deficient (mt) Bad-luciferase construct together with (+) or without (−) DN-ERK5. The following day the cells were cultured in differentiating medium containing NGF (+) for a further 36 h. Where indicated, NGF was removed (−) 18 h before the cells being harvested. Extracts were analyzed for the expression and the phosphorylation (P) of RSK and of CREB by immunoblot (a, b). Chromatin was immunoprecipitated with an antibody to CREB or to an irrelevant protein (Ctrl) to monitor the nonspecific binding to the beads. The precipitated DNA was amplified by semiquantitative PCR (c) or by RT-PCR (d). Input DNA levels were used to monitor transfection efficiency. The data correspond to the mean±S.E. of three independent experiments and are normalized to input DNA levels. Asterisk (*) indicates a non specific band
Chromatin immunoprecipitation (ChIP) analysis using an antibody to CREB demonstrated that CREB interacted with the bad promoter in differentiated PC6.3 cells incubated with NGF (Figure 5c and d). The DNA purified from the cells was amplified by semiquantitative (Figure 5c) or by quantitative RT (Figure 5d) PCR. The amount of PCR product was reduced to that of nonspecific binding following expression of DN-ERK5, NGF withdrawal or mutation of the putative CRE sites. Together, these data indicate that ERK5 suppresses Bad expression by stimulating the transcriptional activity of CREB by RSK and by promoting the binding of CREB to the CRE sites in the bad promoter.
The phosphorylation of Foxo3a by PKB sequesters Foxo3a in the cytoplasm and this prevents increased Bim expression.6, 25 Here we found that absence of ERK5 in SCG neurons (Figure 6a) or ectopic expression of DN-ERK5 in differentiated PC6.3 (Figure 6b) specifically prevented the phosphorylation of PKB at Ser473, but not at Thr308. On the basis of evidence that maximal activation of PKB requires dual phosphorylation at Thr308 and Ser473,26 this result indicates that the level of PKB activity is lower in neurons lacking a functional ERK5 pathway. Consistently, ChIP analysis using an antibody to Foxo3a demonstrated that Foxo3a interacted with the bim promoter in differentiated PC6.3 cells expressing DN-ERK5 (Figure 6c). Together, these studies suggest that ERK5 downregulates Bim expression in neurons by a mechanism that implicates PKB-dependent phosphorylation of Foxo3.
ERK5 regulates Foxo3a-dependent transcription of bim by PKB. (a) Homozygous flox SCG neurons were infected with an adenovirus encoding GFP or Cre. The cells were cultured in presence of NGF (50 ng/ml) for the indicated times. (b, c) PC6.3 cells were transiently transfected with (+) or without (−) DN-ERK5. The following day the cells were cultured in differentiating medium containing NGF for a further 36 h. Extracts were analyzed for the expression and the phosphorylation (P) of PKB at Thr308 or at Ser473 by immunoblot (a, b). Chromatin was immunoprecipitated with an antibody to Foxo3a. The precipitated DNA was amplified by semiquantitative PCR (c). Input DNA levels were used to monitor transfection efficiency. Similar results were obtained in two independent experiments
Discussion
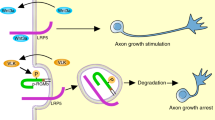
This study provides genetic evidence that ERK5 mediates the survival response of developing sympathetic neurons to NGF by suppressing the transcription of both the bim and bad genes (Figure 7). Our hypothesis that ERK5 prevents Bim expression by inhibiting Foxo3a is supported by the demonstration that Foxo transcription factors activate the bim promoter in sympathetic neurons deprived of NGF.6 Our previous study showed that decreased Foxo3a activity in erk5−/− fibroblasts correlated with a reduced PKB activity compared to wild-type cells.24 Here, we found that the phophorylation of PKB at Ser473, but not at Thr308, was impaired in neurons 36 h after the loss of ERK5. Together with recent evidence that the phophorylation of PKB at Ser473 is required for PKB to phosphorylate Foxo3a,27 this study indicates that ERK5 downregulates Bim expression by promoting PKB-dependent inhibition of Foxo3a (Figure 7). It is interesting to note that the inhibition of PKB signaling by wortmannin further increased the level of neuronal death caused by the absence of ERK5. This is consistent with the idea that defective Ser473 phosphorylation affects only a subset of PKB substrates in vivo.27
Regulation of neuronal survival by the ERK5 cascade. The requirement of ERK5 to phosphorylate PKB at Ser473 prevents the nuclear translocation of the proapoptotic transcription factor Foxo3a and thereby inhibits Bim expression. ERK5 also mediates the phosphorylation of RSK in response to NGF. In the nucleus, RSK stimulates the transcriptional activity of CREB. CREB is a prosurvival transcription factor that can inhibit the transcription of genes responsible for apoptosis including Bad. The binding of CREB to the CRE sites in the bad promoter is dependent on ERK5
Like Bim, we found that increased Bad expression was critical to trigger neuronal apoptosis following the loss of ERK5. Consistent with our data, a previous study has shown that overexpression of Bad in sympathetic neurons overcomes the survival effect of NGF.28 However, the requirement of Bad to mediate the apoptotic response of neurons caused by the loss of ERK5 appears inconsistent with the functional redundancy of Bad with other BH3-only proteins in NGF withdrawal-induced neuronal death.4 This discrepancy can be explained by the fact that, in addition to ERK5, NGF deprivation inhibits ERK1/2 and PKB activity, and stimulates the JNK signaling pathway.16 This leads to additional transcriptional and posttranslational modifications of members of the Bcl-2 family, which sensitize the cells to apoptotic death. For example, NGF-increased Bcl-2 expression in PC12 cells is blocked following inhibition of ERK1/2 signaling.29 In addition, upon phosphorylation by JNK, Bim dissociates from the microtubule-associated dynein motor complex and translocates to the mitochondria.30, 29 Thus, mitochondrial translocation of Bim in NGF-deprived neurons exhibiting a low level of Bcl-2 may allow the activation of Bax independently of Bad. This suggests that the transcriptional upregulation of the bim gene may not be sufficient to trigger neuronal death. Consistent with this, we found that elevated Bim expression in SCG neurons lacking ERK5 and in which the level of Bad is downregulated, is not toxic to the cells. In conclusion, our results support the idea that Bad and Bim are nonredundant BH3-only proteins unless they are posttranslationally modified to increase their proapoptotic function.
The mechanism underlying the transcriptional regulation of bad by ERK5 implicates CREB (Figure 7). This result is strengthened by the finding that CREB binds to the putative Cre sites in the bad promoter. CREB activity is regulated by two potential mechanisms. The first is by phosphorylation at Ser133, which increases the transcriptional activity of CREB.9 Evidence that the phosphorylation of CREB and of RSK was impaired in the absence of ERK5 indicates that ERK5 represses bad expression by RSK-dependent activation of CREB. This model is supported by the finding that RSK is a substrate of ERK510 and that ERK5 contributes to mediating CREB phosphorylation following neurotrophin stimulation of sensory neurons.12 Furthermore, the decrease in CREB phosphorylation caused by the loss of ERK5 followed the same transient kinetics as that of the upregulation of the bad transcript with a maximum after 18 h. The observation that the level of Bad remains elevated up until 48 h after Cre infection suggests that additional mechanisms increase the stability of the protein. The compensatory signaling pathway that partially restores CREB phosphorylation 24 h after the deletion of the erk5 gene, allowing repression of the bad transcription to resume, is unlikely to implicate ERK1/2 considering that the activity of the bad promoter is not affected by UO126. The second mechanism is by the regulation of the binding of CREB to DNA by S-nitrosylation of nuclear proteins that associate with CREB target genes, independently of the phosphorylation of CREB at Ser133.31 Our findings suggest that ERK5-induced CREB-DNA binding may constitute a mechanism that triggers CREB to act as a repressor of gene expression.
Although ERK5 is required for mediating the survival of sensory12 and sympathetic (our results) neurons in vitro, mice lacking ERK5 in the brain do not display any obvious developmental defect.32 However, the sympathetic and sensory nervous systems were not specifically examined in the animal model. Therefore, a more thorough phenotypic analysis of the mice lacking ERK5 will be required to firmly conclude on the role of ERK5 during brain development. In contrast, ERK5 was shown to be essential for neural differentiation in Xenopus early embryonic development.33 This discrepancy between Xenopus and mouse models may be explained by the activation of redundant signaling mechanisms in more complex organisms. Although in vitro ERK1/2 and PKB are not able to fully compensate for the loss of ERK5, activation of ERK1/2 and PKB may be sufficient to sustain the survival of neurons lacking ERK5 in mice.
The requirement of PKB to maintain SCG neuronal survival in response to NGF has been widely documented.16 However, in contrast to our results, most studies have found that inhibition of MEK has minimal effects on NGF-dependent neuronal survival.17, 18, 19, 20 One possible explanation for these controversial findings may lie in the difference in the species from which the neurons were prepared (i.e., rats compared to mice) and in the conditions of the cell cultures. This includes the number of days SCG neurons were kept in vitro before experimentation, which can influence the signaling mechanisms in these postmitotic neurons. In addition, whereas other studies have used PD98059,17, 18, 19, 20 we employed UO126 to block ERK1/2 signaling. To confirm our results, we tested the effect of PD0325901, a novel noncompetitive inhibitor of MEK1 with greater potency.21 Like UO126, we found that PD0325901 decreased the survival of SCG neurons incubated with NGF. These conflicting results emphasize the advantage of genetic deletion analyses over the use of selective inhibitors, which exhibit variability in their efficiency to specifically block the transduction of signals.
The additive effect of erk5 gene deletion with inhibition of ERK1/2 and PKB signaling in vitro suggests that ERK5, ERK1/2 and PKB are components of independent pathways, which contribute to the survival of sympathetic neurons by overlapping mechanisms. For example, although suppression of bad mRNA expression is specifically controlled by ERK5, bim can be transcriptionally regulated by ERK5, ERK1/2 and PKB. In addition, ERK1/2 may be required to maintain Bcl-2 levels,34 and PKB may block the proapoptotic function of Bad by phosphorylation.28, 35 The relative importance of these different survival mechanisms to prevent neuronal death is likely to vary depending on the type of stress. For example, ERK1/2, but not PKB, is required to protect SCG neurons against toxic stimuli.16 The importance of ERK5 in preventing sympathetic neuronal death because of injury or toxicity remains to be tested. In particular, it will be interesting to determine whether the regulation of the BH3-only protein PUMA, which has been implicated in apoptosis induced by DNA damage in sympathetic neurons independently of JNK,36, 37 is controlled by ERK5. Furthermore, phenotypic analysis of the mice lacking ERK5 in the brain will be required to determine whether ERK5 be involved in supporting the survival of neurons under certain pathological situations including aging and neurodegenerative diseases.
Materials and Methods
Cell cultures
Sympathetic neurons were obtained from the SCG of wild type, erk5+/loxP and erk5loxP/loxP new born mice (postnatal days 0–2), as previously described.38 In brief, sympathetic ganglia were trypsinized for 30 min at 37°C. Single-cell suspensions were purified by preplating for 30 min twice on collagen. Nonadherent SCG neurons were collected by centrifugation and cultured for 3 days on poly-L-lysine and laminin coated plates in L15 plating medium containing 3% FBS and 50 ng/ml NGF (Alomone Labs), unless indicated otherwise. All mice employed for this study were hosted in a pathogen-free facility at the University Manchester. Use of animals followed Home Office guidelines and received approval by Manchester University's ethical committee.
The PC6.3 subline of the PC12 cell line were cultured on collagen coated plates in differentiating medium (RPMI containing 1% FBS, 2% horse serum and 100 ng/ml NGF) to obtain a neuronal phenotype, as previously described.6
Genotyping of the cells
SCG neurons were incubated overnight at 55°C with proteinase K. The cell lysates were treated with phenol/TE/hydroxyquinolone and the genomic DNA was isolated by precipitation with isopropanol. Genotype determination was performed by PCR using forward (5′-GCTTCTCCCTGTGATGTGAG-3′) and reverse (5′-TGAGCTACGGGCTTTCG-3′) primers. Fragments (1300 and 250 bp) were amplified from the erk5-flox and disrupted allele, respectively.
Viral infections
The adenoviruses were amplified in HEK-293-T cells and the viral solution was purified on CsCl gradients. The viral infectivity was determined on HEK-293-T cells. After 3 days in culture, SCG neurons were infected with recombinant adenovirus at 100 MOI. Where indicated, recombinant lentivirus at 50 MOI (Mission(R) shRNA lentiviral transduction particles; Sigma) was added 1 h later. The cells were cultured in plating medium containing 3% FBS and 50 ng/ml NGF for a further 48 h, unless indicated otherwise.
Immunoblot analysis
Extracts (15–20 μg) were resolved by SDS-PAGE (12, 10 or 8% polyacrylamide gel) and electrophoretically transferred to an Immobilon-P membrane (Millipore Inc.). Where indicated, the cell lysate was incubated with CIP for 2 h at 37°C before being run on SDS-PAGE. The membranes were incubated with 3% nonfat dry milk at 4°C for 30 min and then probed overnight with antibodies to Bad (BD Transduction Laboratories), Bim (Calbiochem), Bid (R&D Systems), CREB (Cell Signaling Technology), P-CREB at Ser133 (Cell Signaling Technology), ERK1/2 (Santa Cruz), P-ERK1/2 at Thr202 and Tyr204 (Cell Signaling Technology), ERK5 (Upstate Biotechnology), P-ERK5 at Thr218 and Tyr220 (Biosource), JNK (BD Pharmingen), P-JNK at Thr183 and Tyr185 (Cell Signaling Technology), PKB (Cell Signaling Technology), P-PKB at Thr308 or Ser473 (Cell Signaling Technology), RSK (Cell Signaling Technology), phospho-RSK at Thr359 and Ser363 (Cell Signaling Technology), tubulin (Sigma). Immunecomplexes were detected by enhanced chemiluminescence with antimouse or antirabbit immunoglobulin G coupled to horseradish peroxidase as the secondary antibody (Amersham-Pharmacia).
Immunofluorescence
The cells were fixed in methanol before being incubated with specific antibodies to Cre (Chemicon), to Bad (1 : 100; BD Transduction Laboratories) or to the mHsp70 (1 : 200; Cell Signaling Technology). Immune complexes were detected with secondary antimouse and antirabbit antibodies conjugated to Texas red (1 : 500; Invitrogen) or fluorescein (1 : 500; Jackson ImmunoResearch), respectively. Nuclei were stained with DAPI (5 μg/ml). Fluorescence images were viewed with an Olympus Widefield microscope.
Plasmid constructs
Wild-type and double mutant Bim-luciferase plasmids were provided by J. Ham.6 Wild-type and mutant ERK5 were described before.39 The Bad-luciferase construct was created by subcloning fragment −3599 to −1031 of the murine bad promoter into pGL3-basic vector (Promega) using XhoI and HindIII. A mutant (mt) Bad-luciferase construct carrying deletion of two putative CRE-binding sites at −2291 and at −2714 was generated by overlapping PCR.
Reporter gene expression assay
SCG neurons or PC6.3 cells were transiently transfected using the Metafectene™ reagent (Biontex) with the Bad-, Bim- or CRE-luciferase reporter plasmids together with or without an expression vector encoding DN-ERK5. A pRL-Tk plasmid encoding Renilla luciferase was employed for monitoring transfection efficiency. Aliquots of cell lysates were assayed for firefly and Renilla luciferase activities according to the manufacturer's instructions (Promega).
Apoptosis assays
SCG neurons were incubated with Hoechst 33342 (5 μg/ml) and propidium iodide (5 μg/ml) to distinguish viable, necrotic and apoptotic cells. Between 100 and 200 neurons per conditions were scored for apoptosis using a Leica DMIL microscope (Leica, Wetzlar, Germany). Only neurons that had clearly segmented and condensed chromatin were counted as apoptotic. For caspase assays cells were lysed in 20 mM HEPES pH 7.5, 100 mM NaCl, 10 mM DTT, 1 mM EDTA containing 0.1% CHAPS and 10% sucrose. Extracts (5–10 μg) were incubated with 200 μM DEVD-AMC caspase-3 specific fluorogenic substrate for 1 h. Cleavage of the substrate was measured by spectrofluorometer. LDH activity was measured using the CytoTox 96® NonRadioactive Cytotoxicity Assay for Promega. Briefly, a 50 μl aliquot was removed from the cell medium and incubated with LDH substrate for 30 min at room temperature in the dark. Stop solution was added to terminate the reaction. The presence of metabolized substrate was measured at absorbance 490 nm. The amount of LDH detected was proportional to the amount of lysed cells.
Quantitative RT-PCR
Total RNA was isolated using the Trizol™ reagent and cDNA synthesis was carried out as previously described.24 RT quantitative PCRs were performed using the SYBR Green I Core kit (Eurogentec). Primers used were: forward primer, 5′-CTGTGTTCTCTGGCACTCCA-3′ and reverse primer, 5′-TCAGCCACACCCATATCAAA-3′ for erk5; forward primer, 5′-GCCCCTACCTCCCTACAGAC-3′ and reverse primer 5′-AGGACTTGGGGTTTGTGTTG-3′ for bim; forward primer, 5′-CTCCACATCCCGGAACTCTA-3′ and reverse primer 5′-TTAAAGGGACACAGCGATCC-3′ for bad; and forward primer, 5′-CCAACTTGATGTATGAAGGCTTTG-3′ and reverse primer 5′-AATTGGTCTCAAGTCAGTGTACAGGC-3′ for β-actin to generate amplicons of 100, 84, 107 and 91 bp, respectively. PCR products were detected in the ABI-Prism 7700 sequence detection systems (Applied Biosystems). Results were analyzed using the 2−ΔΔG methods. The level of expression of mRNA was normalized to β-actin mRNA.
Chromatin immunoprecipitation assay
PC6.3 cells were transfected with Bad- or Bim-luciferase constructs and subjected to ChIP assay (Active Motif) using a CREB antibody (Santa Cruz), a Foxo3a antibody (Upstate Biotechnology) or a control antibody (AKT; Cell Signaling). Purified immunoprecipitated DNA was subjected to either RT-PCR (using the SYBR Green I Core kit; Eurogentec) or semiquantitative PCR using the following primer sets: promoter region of bad 5′-TTCCTGAGTGGGCCTCATTCCAGCTG-3′ and 5′-CTGTCCTTACACAGTGCCTTC-3′; promoter region of bim 5′-TGCCACCAAAGATCTCTACC-3′ and 5′-GCATTTCCTCACAGAGTTGG-3′. Input DNA levels served as loading controls for transfection efficiency.
Statistical analyses
All P-values were generated using one- or two-way ANOVA analysis, except the data presented in Figure 3a, c and d, where a paired t-test was employed. Ranges were given when the data were obtained from two independent experiments (Figures 2d and 3f, Supplementary Figure S1).
Abbreviations
- CREB:
-
Ca2+/cAMP response element-binding protein
- ERK:
-
extracellular signal-regulated protein kinase
- Foxo3a:
-
Forkhead box O3a
- GFP:
-
green fluorescence protein
- MAPK:
-
mitogen-activated protein kinase
- MEK:
-
MAPK/ERK kinase
- NGF:
-
nerve growth factor
- PKB:
-
protein kinase B
- RSK:
-
p90 ribosomal S6 kinase
- SCG:
-
superior cervical ganglion
References
Oppenheim RW . Cell death during development of the nervous system. Annu Rev Neurosci 1991; 14: 453–501.
Ham JH, Yoon CG, Jung KW, Jang JH . BH3-only proteins: key regulators of neuronal apoptosis. Cell Death Differ 2005; 12: 1015–1020.
Deckwerth TL, Elliott JL, Knudson CM, Johnson Jr EM, Snider WD, Korsmeyer SJ . Bax is required for neuronal death after trophic factor deprivation and during development. Neuron 1996; 17: 401–411.
Putcha GV, Harris CA, Moulder KL, Easton RM, Thompson CB, Johnson EM . Intrinsic and extrinsic pathway signalling during neuronal apoptosis: lessons from the analysis of mutant mice. J Cell Biol 2002; 157: 441–453.
Putcha GV, Moulder KL, Golden JP, Bouillet P, Adams JA, Strasser A et al. Induction of BIM, a pro-apoptotic BH3-only Bcl2 family member, is critical for neuronal apoptosis. Neuron 2001; 29: 615–628.
Gilley J, Coffer PJ, Ham J . FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol 2003; 162: 613–622.
Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J . Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron 2001; 29: 629–643.
Lonze BE, Riccio A, Cohen S, Ginty DD . Apoptosis, axonal growth defects and degeneration of peripheral neurons in mice lacking CREB. Neuron 2002; 34: 371–385.
Lonze BE, Ginty DD . Function and regulation of CREB family transcription factors in the nervous system. Neuron 2002; 35: 605–2334.
Ranganathan A, Pearson G, Chrestensen C, Sturgill T, Cobb M . The MAP kinase ERK5 binds to and phosphorylates p90 RSK. Arch Biochem Biophys 2006; 449: 8–16.
Xing J, Ginty DD, Greenberg ME . Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth-factor regulated CREB kinase. Science 1996; 273: 959–963.
Watson FL, Heerssen HM, Bhattacharyya A, Klesse L, Lin MZ, Segal RA . Neurotrophins use the Erk5 pathway to mediate a retrograde survival response. Nat Neurosci 2001; 4: 981–988.
Liu L, Cavanaugh JE, Wang Y, Sakagami H, Mao Z, Xia Z . ERK5 activation of MEF2-mediated gene expression plays a critical role in BDNF-promoted survival of developing but not mature cortical neurones. Proc Natl Acad Sci USA 2003; 100: 8532–8537.
Shalizi A, Lehtinen M, Gaudilliere B, Donovan N, Han J, Konishi Y et al. Characterization of a neurotrophin signaling mechanism that mediates neuron survival in a temporally specific pattern. J Neurosci 2003; 23: 7326–7336.
Wang X, Tournier C . Regulation of cellular functions by the ERK5 signalling pathway. Cell Signal 2006; 18: 753–760.
Kaplan DR, Miller FD . Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol 2000; 10: 381–391.
Mazzoni IE, Said FA, Aloyz R, Miller FD, Kaplan D . Ras regulates sympathetic neuron survival by suppressing the p53-mediated cell death pathway. J Neurosci 1999; 19: 9716–9727.
Creedon DJ, Johnson EM, Lawrence JC . Mitogen activated protein kinase-independent pathways mediate the effects of nerve growth factor and cAMP on neuronal survival. J Biol Chem 1996; 271: 20713–20718.
Klesse LJ, Parada LF . p21 Ras and phosphatidylinositol-3 kinase are required for survival of wild-type and NF1 mutant sensory neurons. J Neurosci 1998; 18: 10420–10428.
Virdee K, Tolkovsky AM . Inhibition of p42 and p44 mitogen-activated protein kinase activity by PD98059 does not suppress nerve growth factor-induced survival of sympathetic neurones. J Neurochem 1996; 67: 1801–1805.
Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, Mclauchlan H et al. The selectivity of protein kinase inhibitors: a further update. Biochem J 2007; 408: 297–315.
Pi X, Yan C, Berk BC . Big mitogen-activated protein kinase (BMK1)/ERK5 protects endothelial cells from apoptosis. Circ Res 2004; 94: 362–369.
Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ . Bad, a heterodimeric partner for Bcl-Xl and Bcl-2, displaces Bax and promotes cell death. Cell 1995; 80: 285–291.
Wang X, Finegan KG, Robinson AC, Knowles L, Khosravi-Far R, Hinchliffe KA et al. Activation of extracellular signal-regulated protein kinase 5 down-regulates FasL upon osmotic stress. Cell Death Differ 2006; 13: 2099–2108.
Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999; 96: 857–868.
Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J 1996; 15: 6541–6551.
Jacinto E, Facchinetti V, Liu D, Soto S, Wei S, Jung SY et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 2006; 127: 125–137.
Roberts ML, Virdee K, Sampson CPB, Gordon I, Parone P, Tolkovsky A . The combination of Bcl-2 expression and NGF-deprivation facilitates the selective destruction of the Bad protein in living sympathetic neurons. Mol Cell Neurosci 2000; 16: 97–110.
Putcha GV, Le S, Frank S, Besirli CG, Clark K, Chu B et al. JNK-mediated BIM phosphorylation potentiates BAX-dependent apoptosis. Neuron 2003; 38: 899–914.
Lei K, Davis RJ . JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc Natl Acad Sci USA 2003; 100: 2432–2437.
Riccio A, Alvania RS, Lonze RE, Ramanan N, Kim T, Huang Y et al. A nitric oxide signalling pathway controls CREB-mediated gene expression in neurones. Mol Cell 2006; 21: 283–294.
Hayashi M, Lee J-D . Role of the BMK1/ERK5 signaling pathway: lessons from knockout mice. J Mol Med 2004; 82: 800–808.
Nishimoto S, Kusakabe M, Nishida E . Requirement of the MEK5–ERK5 pathway for neural differentiation in Xenopus embryonic development. EMBO Rep 2005; 6: 1064–1069.
Liu Y-Z, Boxer LM, Latchman DS . Activation of the Bcl-2 promoter by nerve growth factor is mediated by the p42/p44 MAPK cascade. Nucleic Acids Res 1999; 27: 2086–2090.
Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 1997; 91: 231–241.
Anderson CNG, Tolkovsky AM . A role for MAPK/ERK in sympathetic neuron survival: protection against a p53-dependent, JNK-independent induction of apoptosis by cytosine arabinoside. J Neurosci 1999; 19: 664–673.
Wyttenbach A, Tolkovsky AM . The BH3-only protein Puma is both necessary and sufficient for neuronal apoptosis induced by DNA damage in sympathetic neurons. J Neurochem 2006; 96: 1213–1226.
Whitfield J, Neame SJ, Ham J . Methods for culturing primary sympathetic neurons and for determining neuronal viability. Methods Mol Biol 2004; 282: 157–168.
Seyfried J, Wang X, Kharebava G, Tournier C . A novel MAPK docking site in the N-terminus of MEK5α organizes the components of the ERK5 signaling pathway. Mol Cell Biol 2005; 25: 9820–9828.
Acknowledgements
We are indebted to B Carter and A Tolkovsky for their generosity in providing recombinant adenoviruses. We thank J Ham for providing us with the PC6.3 cells and the Bim-luciferase constructs and A Whitmarsh for critically reviewing the article. PD0325901 was a gift from P Cohen and we thank N Shpiro for synthesizing the compound. This work was supported by the BBSRC, the Wellcome Trust and a Lister Institute of Preventive Medicine Research Fellowship to CT.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by D Rubinsztein
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Finegan, K., Wang, X., Lee, EJ. et al. Regulation of neuronal survival by the extracellular signal-regulated protein kinase 5. Cell Death Differ 16, 674–683 (2009). https://doi.org/10.1038/cdd.2008.193
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.193
Keywords
This article is cited by
-
Newt A1 cell-derived extracellular vesicles promote mammalian nerve growth
Scientific Reports (2023)
-
Autophagy and apoptosis cascade: which is more prominent in neuronal death?
Cellular and Molecular Life Sciences (2021)
-
Neurotrophin Signaling and Stem Cells—Implications for Neurodegenerative Diseases and Stem Cell Therapy
Molecular Neurobiology (2017)
-
The MEK-ERK pathway negatively regulates bim expression through the 3' UTR in sympathetic neurons
BMC Neuroscience (2011)
-
Spatial cycles in G-protein crowd control
The EMBO Journal (2010)