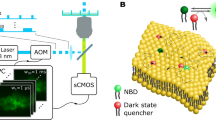
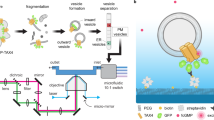
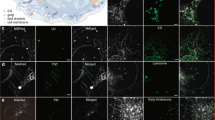
Abstract
Cholesterol-mediated lipid interactions are thought to have a functional role in many membrane-associated processes such as signalling events1,2,3,4,5. Although several experiments indicate their existence, lipid nanodomains (‘rafts’) remain controversial owing to the lack of suitable detection techniques in living cells4,6,7,8,9. The controversy is reflected in their putative size of 5–200 nm, spanning the range between the extent of a protein complex and the resolution limit of optical microscopy. Here we demonstrate the ability of stimulated emission depletion (STED) far-field fluorescence nanoscopy10 to detect single diffusing (lipid) molecules in nanosized areas in the plasma membrane of living cells. Tuning of the probed area to spot sizes ∼70-fold below the diffraction barrier reveals that unlike phosphoglycerolipids, sphingolipids and glycosylphosphatidylinositol-anchored proteins are transiently (∼10–20 ms) trapped in cholesterol-mediated molecular complexes dwelling within <20-nm diameter areas. The non-invasive optical recording of molecular time traces and fluctuation data in tunable nanoscale domains is a powerful new approach to study the dynamics of biomolecules in living cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Simons, K. & Ikonen, E. Functional rafts in cell membranes. Nature 387, 569–572 (1997)
Brown, D. A. & London, E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275, 17221–17224 (2000)
Fielding, C. J. Lipid Rafts and Caveolae (Wiley-VCH, 2006)
Jacobson, K., Mouritsen, O. G. & Anderson, G. W. Lipid rafts: at a crossroad between cell biology and physics. Nature Cell Biol. 9, 7–14 (2007)
Hanzal-Bayer, M. F. & Hancock, J. F. Lipid rafts and membrane traffic. FEBS Lett. 581, 2098–2104 (2007)
Munro, S. Lipid rafts: elusive or illusive? Cell 115, 377–388 (2003)
Lommerse, P. H. M., Spaink, H. P. & Schmidt, T. In vivo plasma membrane organization: results of biophysical approaches. Biochim. Biophys. Acta 1664, 119–131 (2004)
Hancock, J. F. Lipid rafts: contentious only from simplistic standpoints. Nature Rev. Mol. Cell Biol. 7, 456–462 (2006)
Shaw, A. S. Lipid rafts: now you see them, now you don’t. Nature Immunol. 7, 1139–1142 (2006)
Hell, S. W. & Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: stimulated emission depletion microscopy. Opt. Lett. 19, 780–782 (1994)
Pike, L. J. Rafts defined: a report on the Keystone symposium on lipid rafts and cell function. J. Lipid Res. 47, 1597–1598 (2006)
Fujita, A. et al. Gangliosides GM1 and GM3 in the living cell membrane form clusters susceptible to cholesterol depletion and chilling. Mol. Biol. Cell 18, 2112–2122 (2007)
Binnig, G., Quate, C. F. & Gerber, C. Atomic force microscope. Phys. Rev. Lett. 56, 930–933 (1986)
Pohl, D. W., Denk, W. & Lanz, M. Optical stethoscopy: Image recording with resolution λ/20. Appl. Phys. Lett. 44, 651–653 (1984)
Zacharias, D. A., Violin, J. D., Newton, A. C. & Tsien, R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 (2002)
Saxton, M. J. & Jacobson, K. Single particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26, 373–399 (1997)
Schütz, G. J., Kada, G., Pastushenko, V. P. & Schindler, H. Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. EMBO J. 19, 892–901 (2000)
Fujiwara, T., Ritchie, K., Murakoshi, H., Jacobson, K. & Kusumi, A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157, 1071–1081 (2002)
Yechiel, E. & Edidin, M. Micrometer-scale domains in fibroblast plasma membranes. J. Cell Biol. 105, 755–760 (1987)
Feder, T. J., Brust-Mascher, I., Slattery, J. P., Baird, B. A. & Webb, W. W. Constrainted diffusion or immobile fraction on cell surfaces: a new interpretation. Biophys. J. 70, 2767–2773 (1996)
Fahey, P. F. et al. Lateral diffusion in planar lipid bilayers. Science 195, 305–306 (1977)
Schwille, P., Korlach, J. & Webb, W. W. Fluorescence correlation spectroscopy with single-molecule sensitivity on cell and model membranes. Cytometry 36, 176–182 (1999)
Wawrezinieck, L., Rigneault, H., Marguet, D. & Lenne, P.-F. Fluorescence correlation spectroscopy: diffusion laws to probe the submicron cell membrane organization. Biophys. J. 89, 4029–4042 (2005)
Wenger, J. et al. Diffusion analysis within single nanometric apertures reveals the ultrafine cell membrane organization. Biophys. J. 92, 913–919 (2007)
Willig, K. I., Rizzoli, S. O., Westphal, V., Jahn, R. & Hell, S. W. STED-microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature 440, 935–939 (2006)
Hell, S. W. Far-field optical nanoscopy. Science 316, 1153–1158 (2007)
Magde, D., Elson, E. L. & Webb, W. W. Thermodynamic fluctuations in a reacting system - measurement by fluorescence correlation spectroscopy. Phys. Rev. Lett. 29, 705–708 (1972)
Osborn, M., Franke, W. W. & Weber, K. Visualization of a system of filaments 7–10 nm thick in cultured cells of an epithelioid line (PtK2) by immunofluorescence microscopy. Proc. Natl Acad. Sci. USA 74, 2490–2494 (1977)
Martin, O. C. & Pagano, R. C. Internalization and sorting of a fluorescent analogue of glucosylceramide to the Golgi apparatus of human skin fibroblasts: utilization of endocytic and nonendocytic transport mechanisms. J. Cell Biol. 125, 769–781 (1994)
Schwarzmann, G., Hofmann, P., Pütz, U. & Albrecht, B. Demonstration of direct glycosylation of nondegradable glucosylceramide analogs in cultured cells. J. Biol. Chem. 270, 21271–21276 (1995)
Keller, J., Schönle, A. & Hell, S. W. Efficient fluorescence inhibition patterns for RESOLFT microscopy. Opt. Express 15, 3361–3371 (2007)
Willig, K. I. et al. Nanoscale resolution in GFP-based microscopy. Nature Methods 3, 721–723 (2006)
Acknowledgements
We thank J. Jethwa, B. Rankin and M. Hilbert for critical reading, K. Willig for help with the setup, T. Lang, R. Wagner and H. Rigneault for valuable discussions, R. Machinek and H. Frauendorf for recording the NMR and mass spectra, and S. Yan for help with the synthesis.
Author Contributions C.R. and B.H. performed experiments, C.E. and C.R. analysed data, R.M. prepared samples and performed washing experiments, G.S., K.S., S.P. and V.N.B. synthesized fluorescently labelled lipids and performed chromatography, C.v.M., A.S., C.R. and C.E. realized and analysed simulated data, C.E. and S.W.H. designed experiments and wrote the paper. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Materials and Methods , Supplementary Data, Supplementary Figures 1-11 with Legends and Supplementary References (PDF 1283 kb)
Rights and permissions
About this article
Cite this article
Eggeling, C., Ringemann, C., Medda, R. et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1162 (2009). https://doi.org/10.1038/nature07596
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07596
This article is cited by
-
Membrane organization by tetraspanins and galectins shapes lymphocyte function
Nature Reviews Immunology (2024)
-
Current capabilities and future perspectives of FCS: super-resolution microscopy, machine learning, and in vivo applications
Communications Biology (2023)
-
Regulation of membrane protein structure and function by their lipid nano-environment
Nature Reviews Molecular Cell Biology (2023)
-
Single-photon microscopy to study biomolecular condensates
Nature Communications (2023)
-
Direct regulation of the voltage sensor of HCN channels by membrane lipid compartmentalization
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.