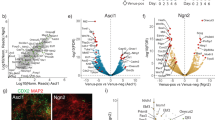
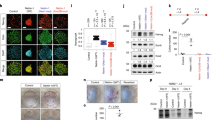
Abstract
The neural fate is generally considered to be the intrinsic direction of embryonic stem (ES) cell differentiation. However, little is known about the intracellular mechanism that leads undifferentiated cells to adopt the neural fate in the absence of extrinsic inductive signals. Here we show that the zinc-finger nuclear protein Zfp521 is essential and sufficient for driving the intrinsic neural differentiation of mouse ES cells. In the absence of the neural differentiation inhibitor BMP4, strong Zfp521 expression is intrinsically induced in differentiating ES cells. Forced expression of Zfp521 enables the neural conversion of ES cells even in the presence of BMP4. Conversely, in differentiation culture, Zfp521-depleted ES cells do not undergo neural conversion but tend to halt at the epiblast state. Zfp521 directly activates early neural genes by working with the co-activator p300. Thus, the transition of ES cell differentiation from the epiblast state into neuroectodermal progenitors specifically depends on the cell-intrinsic expression and activator function of Zfp521.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Muñoz-Sanjuán, I. & Hemmati-Brivanlou, A. Neural induction, the default model and embryonic stem cells. Nature Rev. Neurosci. 3, 271–280 (2002)
De Robertis, E. M. & Sasai, Y. A common plan for dorsoventral patterning in Bilateria. Nature 380, 37–40 (1996)
Sasai, Y., Lu, B., Steinbeisser, H. & De Robertis, E. M. Regulation of neural induction by the chd and BMP-4 antagonistic patterning signals in Xenopus. Nature 376, 333–336 (1995)
Lamb, T. M. et al. Neural induction by the secreted polypeptide noggin. Science 262, 713–718 (1993)
Reversade, B., Kuroda, H., Lee, H., Mays, A. & De Robertis, E. M. Depletion of Bmp2, Bmp4, Bmp7 and Spemann organizer signals induces massive brain formation in Xenopus embryos. Development 132, 3381–3392 (2005)
Di Giorgio, F. P., Carrasco, M. A., Siao, M. C., Maniatis, T. & Eggan, K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nature Neurosci. 10, 608–614 (2007)
Kawasaki, H. et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron 28, 31–40 (2000)
Watanabe, K. et al. Directed differentiation of telencephalic precursors from embryonic stem cells. Nature Neurosci. 8, 288–296 (2005)
Smukler, S. R., Runciman, S. B., Xu, S. & van der Kooy, D. Embryonic stem cells assume a primitive neural stem cell fate in the absence of extrinsic influences. J. Cell Biol. 172, 79–90 (2006)
Wichterle, H., Lieberam, I., Porter, J. A. & Jessell, T. M. Directed differentiation of embryonic stem cells into motor neurons. Cell 110, 385–397 (2002)
Abranches, E. et al. Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PLoS ONE 4, e6286 (2009)
Eiraku, M. et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532 (2008)
Aubert, J. et al. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-GFP knock-in mice. Proc. Natl Acad. Sci. USA 100, 11836–11841 (2003)
Jakt, L. M., Okada, M. & Nishikawa, S.-I. An open source client-server system for the analysis of Affymetrix microarray data. Genome Informat. 14, 276–277 (2003)
Warming, S. et al. Evi3, a common retroviral integration site in murine B-cell lymphoma, encodes an EBFAZ-related Krüppel-like zinc finger protein. Blood 101, 1934–1940 (2003)
Bond, H. M. et al. Early hematopoietic zinc finger protein-zinc finger protein 521: a candidate regulator of diverse immature cells. Int. J. Biochem. Cell Biol. 40, 848–854 (2008)
von Bubnoff, A. et al. Phylogenetic footprinting and genome scanning identify vertebrate BMP response elements and new target genes. Dev. Biol. 281, 210–226 (2005)
Benchabane, H. & Wrana, J. L. GATA- and Smad1-dependent enhancers in the Smad7 gene differentially interpret bone morphogenetic protein concentrations. Mol. Cell. Biol. 23, 6646–6661 (2003)
Gajović, S., St-Onge, L., Yokota, Y. & Gruss, P. Retinoic acid mediates Pax6 expression during in vitro differentiation of embryonic stem cells. Differentiation 62, 187–192 (1997)
Takada, S. et al. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 8, 174–189 (1994)
Kunath, T. et al. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134, 2895–2902 (2007)
Tesar, P. J. et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 (2007)
Ding, J. et al. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature 395, 702–707 (1998)
Lin, A. C., Roche, A. E., Wilk, J. & Svensson, E. C. The N termini of Friend of GATA (FOG) proteins define a novel transcriptional repression motif and a superfamily of transcriptional repressors. J. Biol. Chem. 279, 55017–55023 (2004)
Wu, M. et al. Zfp521 antagonizes Runx2, delays osteoblast differentiation in vitro, and promotes bone formation in vivo. Bone 44, 528–536 (2009)
Chan, H. M. & La Thangue, N. B. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114, 2363–2373 (2001)
Visel, A. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (2009)
Brunelli, S., Silva Casey, E., Bell, D., Harland, R. & Lovell-Badge, R. Expression of Sox3 throughout the developing central nervous system is dependent on the combined action of discrete, evolutionarily conserved regulatory elements. Genesis 36, 12–24 (2003)
de Ruijter, A. J., van Gennip, A. H., Caron, H. N., Kemp, S. & van Kuilenburg, A. B. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749 (2003)
Tsai, R. Y. & Reed, R. R. Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol. Cell. Biol. 18, 6447–6456 (1998)
Zhao, S., Nichols, J., Smith, A. G. & Li, M. SoxB transcription factors specify neuroectodermal lineage choice in ES cells. Mol. Cell. Neurosci. 27, 332–342 (2004)
Kuhlbrodt, K. et al. Cooperative function of POU proteins and SOX proteins in glial cells. J. Biol. Chem. 273, 16050–16057 (1998)
Tanaka, S. et al. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol. Cell. Biol. 24, 8834–8846 (2004)
Zwart, R., Broos, L., Grosveld, G. & Meijer, D. The restricted expression pattern of the POU factor Oct-6 during early development of the mouse nervous system. Mech. Dev. 54, 185–194 (1996)
Jørgensen, H. F. et al. REST selectively represses a subset of RE1-containing neuronal genes in mouse embryonic stem cells. Development 136, 715–721 (2009)
Watanabe, K. et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnol. 25, 681–686 (2007)
Nagy, A., Gertsenstein, M. G., Vintersten, K. & Behringer, R. Manipulating the Mouse Embryo (Cold Spring Harbor Press, 2003)
Ikeya, M. et al. Gene disruption/knock-in analysis of mONT3: vector construction by employing both in vivo and in vitro recombinations. Int. J. Dev. Biol. 49, 807–823 (2005)
Acknowledgements
We are grateful to H. Niwa for comments; M. Okada for advice on the GeneChip study; Y. Toyooka-Kamiya for constant encouragement and technical advice; H. Akimaru for advice on the shRNA knockdown study; and members of the Sasai laboratory for discussion and advice. This work was supported by grants-in-aid from MEXT, the Kobe Cluster Project and the Leading Project (Y.S.).
Author information
Authors and Affiliations
Contributions
Y.S. designed the research and D.K. performed the majority of mES cell experiments with technical help from S.B., M.K. and R.Y. N.S., M.O., H.I. and K.W. collaborated with D.K. in the ChIP, hES cell, Xenopus and SFEB experiments. H.K. and K.N. performed the blastocyst injections and L.M.J. and S.-i.N. helped GeneChip screening.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-12 with legends and Supplementary Tables 1-2. (PDF 0 kb)
Rights and permissions
About this article
Cite this article
Kamiya, D., Banno, S., Sasai, N. et al. Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature 470, 503–509 (2011). https://doi.org/10.1038/nature09726
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09726
This article is cited by
-
Cooperative regulation of Zhx1 and hnRNPA1 drives the cardiac progenitor-specific transcriptional activation during cardiomyocyte differentiation
Cell Death Discovery (2023)
-
Massively parallel reporter perturbation assays uncover temporal regulatory architecture during neural differentiation
Nature Communications (2022)
-
Reprocessing seafood waste: challenge to develop aquatic clean meat from fish cells
npj Science of Food (2022)
-
Signal requirement for cortical potential of transplantable human neuroepithelial stem cells
Nature Communications (2022)
-
An Overview of Mesenchymal Stem Cell-based Therapy Mediated by Noncoding RNAs in the Treatment of Neurodegenerative Diseases
Stem Cell Reviews and Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.