Abstract
During telophase, the nuclear envelope (NE) reforms around daughter nuclei to ensure proper segregation of nuclear and cytoplasmic contents1,2,3,4. NE reformation requires the coating of chromatin by membrane derived from the endoplasmic reticulum, and a subsequent annular fusion step to ensure that the formed envelope is sealed1,2,4,5. How annular fusion is accomplished is unknown, but it is thought to involve the p97 AAA-ATPase complex and bears a topological equivalence to the membrane fusion event that occurs during the abscission phase of cytokinesis1,6. Here we show that the endosomal sorting complex required for transport-III (ESCRT-III) machinery localizes to sites of annular fusion in the forming NE in human cells, and is necessary for proper post-mitotic nucleo-cytoplasmic compartmentalization. The ESCRT-III component charged multivesicular body protein 2A (CHMP2A) is directed to the forming NE through binding to CHMP4B, and provides an activity essential for NE reformation. Localization also requires the p97 complex member ubiquitin fusion and degradation 1 (UFD1). Our results describe a novel role for the ESCRT machinery in cell division and demonstrate a conservation of the machineries involved in topologically equivalent mitotic membrane remodelling events.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Burke, B. The nuclear envelope: filling in gaps. Nature Cell Biol. 3, E273–E274 (2001).
Anderson, D. J. & Hetzer, M. W. Shaping the endoplasmic reticulum into the nuclear envelope. J. Cell Sci. 121, 137–142 (2008).
Schooley, A., Vollmer, B. & Antonin, W. Building a nuclear envelope at the end of mitosis: coordinating membrane reorganization, nuclear pore complex assembly, and chromatin de-condensation. Chromosoma 121, 539–554 (2012).
Burke, B. & Ellenberg, J. Remodelling the walls of the nucleus. Nature Rev. Mol. Cell Biol. 3, 487–497 (2002).
Lu, L., Ladinsky, M. S. & Kirchhausen, T. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J. Cell Biol. 194, 425–440 (2011).
Hetzer, M. et al. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nature Cell Biol. 3, 1086–1091 (2001).
McCullough, J., Colf, L. A. & Sundquist, W. I. Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 82, 663–692 (2013).
Carlton, J. G., Agromayor, M. & Martin-Serrano, J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl Acad. Sci. USA 105, 10541–10546 (2008).
Carlton, J. G. & Martin-Serrano, J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908–1912 (2007).
Elia, N., Sougrat, R., Spurlin, T. A., Hurley, J. H. & Lippincott-Schwartz, J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl Acad. Sci. USA 108, 4846–4851 (2011).
Morita, E. et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26, 4215–4227 (2007).
Jouvenet, N., Zhadina, M., Bieniasz, P. D. & Simon, S. M. Dynamics of ESCRT protein recruitment during retroviral assembly. Nature Cell Biol. 13, 394–401 (2011).
Carlton, J. G., Caballe, A., Agromayor, M., Kloc, M. & Martin-Serrano, J. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science 336, 220–225 (2012).
Clever, M., Funakoshi, T., Mimura, Y., Takagi, M. & Imamoto, N. The nucleoporin ELYS/Mel28 regulates nuclear envelope subdomain formation in HeLa cells. Nucleus 3, 187–199 (2012).
Stauffer, D. R., Howard, T. L., Nyun, T. & Hollenberg, S. M. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J. Cell Sci. 114, 2383–2393 (2001).
Morita, E. et al. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9, 235–242 (2011).
Morita, E. et al. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl Acad. Sci. USA 107, 12889–12894 (2010).
Asencio, C. et al. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell 150, 122–135 (2012).
Buchkovich, N. J., Henne, W. M., Tang, S. & Emr, S. D. Essential N-terminal insertion motif anchors the ESCRT-III filament during MVB vesicle formation. Dev. Cell 27, 201–214 (2013).
Ramadan, K. et al. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature 450, 1258–1262 (2007).
Dobrynin, G. et al. Cdc48/p97-Ufd1-Npl4 antagonizes Aurora B during chromosome segregation in HeLa cells. J. Cell Sci. 124, 1571–1580 (2011).
Ritz, D. et al. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nature Cell Biol. 13, 1116–1123 (2011).
Anderson, D. J. & Hetzer, M. W. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J. Cell Biol. 182, 911–924 (2008).
Sorg, G. & Stamminger, T. Mapping of nuclear localization signals by simultaneous fusion to green fluorescent protein and to β-galactosidase. Biotechniques 26, 858–862 (1999).
Lee, C.-P. et al. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein–Barr Virus. PLoS Pathog. 8, e1002904 (2012).
Pawliczek, T. & Crump, C. M. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 83, 11254–11264 (2009).
Speese, S. D. et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149, 832–846 (2012).
Webster, B. M., Colombi, P., Jäger, J. & Lusk, C. P. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 159, 388–401 (2014).
Agromayor, M. et al. Essential role of hIST1 in cytokinesis. Mol. Biol. Cell 20, 1374–1387 (2009).
Martin-Serrano, J., Yarovoy, A., Perez-Caballero, D., Bieniasz, P. D. & Yaravoy, A. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl Acad. Sci. USA 100, 12414–12419 (2003).
Meerang, M. et al. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nature Cell Biol. 13, 1376–1382 (2011).
van Weering, J. R. T. et al. Intracellular membrane traffic at high resolution. Methods Cell Biol. 96, 619–648 (2010).
Acknowledgements
J.G.C. is a Wellcome Trust Research Career Development Fellow. We acknowledge the Nikon Imaging Centre at KCL and the NIHR Comprehensive Biomedical Research Centre at Guy's and St Thomas’ NHS Foundation Trust for access to core equipment. We thank the staff of the Wolfson Bioimaging Facility for their support. We thank J. Martin-Serrano for gifts of plasmids and cells.
Author information
Authors and Affiliations
Contributions
J.G.C. conceived the study. P.V., L.H. and J.M. designed, performed and analysed electron microscopy experiments. J.G.C. and Y.O. designed, performed and analysed data from other experiments. J.G.C. wrote the manuscript with assistance from all other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Localization of ESCRT components during the cell cycle.
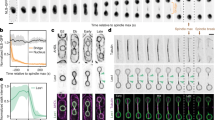
a, b, Immunofluorescence analysis of HeLa cells stained with anti-tubulin, anti-CHMP2A or anti-CHMP2B and DAPI (a). Images in a are representative of two acquired images per field of view. Cells in b were treated with control or CHMP2A-targeting siRNA; images representative of four (control) or two (CHMP2A siRNA) acquired images. c, Deconvolved projections of HeLa cells stained with anti-CHMP2A and DAPI, corresponding to stills from Supplementary Video 1. Images representative of two deconvolved image series. d, HeLa cells stably expressing GFP–CHMP4B were imaged live during the anaphase to telophase transition. Telophase frames at 30-s intervals are presented, corresponding to stills from Supplementary Video 2. Images representative of four acquisitions. e, Immunofluorescence analysis of human diploid fibroblasts stained with anti-CHMP2A, anti-tubulin and DAPI, images representative of three acquired cells per cell cycle phase. f, g, Immunofluorescence analysis of HeLa cells stained with anti-CHMP2A, DAPI and either anti-mAb414 (f) or anti-LaminA/C (g), images representative of five acquired cells. Arrowheads indicate regions of formed nuclear pores or lamina as indicated. h, Quantification of abnormal nuclei (the presence of multiple lobes, micronuclei, lamina ingression or invagination) in HeLa cells transfected with the indicated siRNA and stained with anti-LaminA/C (1,300 cells over 5 experiments quantified per treatment; data are mean ± s.d.). Images representative of three (control, CHMP2A siRNA) or two (LEM4 siRNA) acquired fields of view and resolved cell lysates were examined by western blotting with anti-CHMP2A, anti-CHMP2B or anti-GAPDH antisera as indicated. Scale bars, 10 µm.
Extended Data Figure 2 Correlative light and electron microscopy (CLEM) of endogenous CHMP2A localization in telophase NE.
a–c, Phase-contrast (a), correlative immunofluorescence (b) and transmission electron microscopy of HeLa cells stained with anti-CHMP2A, detected by Alexa-594-fluoronanogold and DAPI. Boxed region in a is shown in b; boxed region in b is shown in c. In all cases, images representative of three cells prepared for CLEM. d, 3D rendering of tomographic reconstruction of forming NE from boxed region in c and Fig. 1d; a single example of a nucleo-cytoplasmic channel was selected for 3D rendering. e–g, Z-slices extracted from tomographic reconstructions of forming NE depicting CHMP2A localization to isolated vesicles (e, i) and nucleo-cytoplasmic channels (arrows in e, ii, f, g) at the indicated Z-heights Localization of CHMP2A to nucleo-cytoplasmic channels was observed in three independent cells; data from a second cell are presented in Extended Data Fig. 3. Note CHMP2A localization to nucleo-cytoplasmic channels is distinct from nuclear pores (asterisk in f). h, Quantification of CHMP2A labelling from two independently prepared cells. Channels were defined as discontinuities up to 80 nm, and gaps were defined as discontinuities over 80 nm. Distances of the gold-particles from channels or gaps were measured on the tomograms in three-dimensions and plotted as a histogram. Most (74.4%) of the gold label was found within 150 nm of nucleo-cytoplasmic channels, and most (70.6%) of the gold label was found more than 150 nm from the larger gaps in the NE. Scale bars, 10 µm (b) and 200 nm (f, g).
Extended Data Figure 3 CLEM of endogenous CHMP2A localization in telophase NE.
a–c, Phase-contrast (a), correlative immunofluorescence (b) and transmission electron microscopy (c) of a second HeLa cell stained with anti-CHMP2A, detected by Alexa-594-fluoronanogold and DAPI. Boxed region in a is shown in b; boxed region in b is shown in c. d, Z-slices extracted from tomographic reconstruction of forming NE from boxed region in c depicting CHMP2A-localization to nucleo-cytoplasmic channels at the indicated Z-heights. Arrow indicates nucleo-cytoplasmic channel. Images in all cases representative of 3 cells processed for CLEM, quantification of gold localization given in Extended Data Fig. 2H. Scale bars, 24 µm (b), 1 µm (c) and 200 nm (d).
Extended Data Figure 4 Mitotic defects in cells reliant on mutated forms of CHMP2A.
a, Quantification of CHMP2A recruitment to the telophase NE or the midbody from Fig. 2c (n = 3, 10 cells (midbody or telophase) scored per experiment). b, Quantification of cytokinetic failure from cells treated with the indicated siRNA (300 cells were quantified per experiment, from three independent experiments). Data are mean ± s.d.
Extended Data Figure 5 Screening for ESCRT–p97 complex interactions.
a–d, β-galactosidase activity of yeast co-transformed with the indicated Gal4 (ESCRT)- and VP16-fused proteins (n = 2). e, Resolved cell lysates and glutathione-bound fractions from 293T cells transfected with the indicated fusion proteins were examined by western blotting with anti-GFP (n = 3). f, β-galactosidase activity of yeast co-transformed with the indicated Gal4- and VP16-fused proteins (n = 3). g, Microscale thermophoresis experiments detailing binding of CHMP2A to GST (n = 4), His–UFD1 (n = 5) or His–UFD1(1–257) (n = 4). As no reduction in thermophoresis signal was observed for GST or His–UFD1(1–257) across the concentration range, we present here the average thermophoresis signal change at equivalent protein concentrations for these three proteins, normalized to zero at the concentration in capillary 1. h, Alexa-647-labelled CHMP2A, His–UFD1 and His–UFD1(1–257) were examined by infrared imaging or Coomassie staining. Data are mean ± s.d.
Extended Data Figure 6 UFD1 depletion does not affect ESCRT-dependent receptor degradation, lentivirus release or cytokinetic abscission.
a, Resolved cell lysates of HeLa cells transfected with the indicated siRNA were examined by western blotting with anti-UFD1 or anti-HSP90 antisera. b, Resolved lysates of human diploid fibroblasts transfected with the indicted siRNA and treated for the indicated times with epidermal growth factor (20 ng ml−1) were examined by western blotting with anti-EGFR, anti-UFD1 and anti-GAPDH antisera. EGFR degradation was quantified by densitometry (n = 3). c, Resolved cell lysates from 293T cells transfected with the indicated HIV-1 based lentiviral plasmids, a virally packaged GFP-plasmid, and the indicated siRNA were examined by western blotting with anti-p24 capsid, -HSP90, -TSG101, -CHMP2A, -CHMP2B and -UFD1 antibodies. Viral supernatants were collected and used to infect target HeLa cells. Resolved virions present in the 293T supernatant were examined by western blotting with anti-p24 capsid. Resolved lysates of infected HeLa cells were examined by western blotting with anti-GFP. Virion release was the ratio of released to cellular p24 capsid, as quantified by densitometry (n = 2); infectivity was quantified as GFP signal in target cells, as quantified by densitometry (n = 2). d, siRNA-transfected HeLa cells were fixed and stained with anti-tubulin. Multinucleate cells (n = 5) or cells connected by midbodies (n = 5) were scored visually, 300 cells scored per experiment. Data are mean ± s.d.
Extended Data Figure 7 ESCRT depletion impairs NE-rim formation.
a, b, Timelapse microscopy analysis and quantification of NE-rim formation in HeLa cells stably expressing YFP–LAP2β and mCh–H2B and treated with the indicated siRNA. Scale bars, 10 µm. Time for rim formation post anaphase onset given (mins) (control, 8.53 ± 0.09, 226 cells analysed over 8 independent experiments; CHMP2A-1, 7.60 ± 0.09, 205 cells analysed over 7 independent experiments; CHMP2A-2, 6.86 ± 0.12, 37 cells analysed over 2 independent experiments; CHMP2B, 6.92 ± 0.09, 79 cells analysed over 4 independent experiments; CHMP2A and CHMP2B, 6.84 ± 0.13, 50 cells analysed over 2 independent experiments; CHMP4B, 7.07 ± 0.14, 44 cells analysed over 2 independent experiments; UFD1, 9.2 ± 0.18, 39 cells analysed over 3 independent experiments). Data are mean ± s.e.m. (in minutes). Images representative of the indicated number of cell analysed. c, Resolved cell lysates from a were analysed by western blotting with the indicated antisera.
Extended Data Figure 8 ESCRT depletion does not impair nuclear pore formation.
a, Schematic of nuclear envelope integrity assay. b, Control-siRNA-treated HeLa cells reporting nucleo-cytoplasmic partitioning using the GFP–NLS–βGal assay, average NE compartmentalization from 20 cells presented. Nucleo-cytoplasmic partitioning stabilizes at 85 min (indicated by arrow). c, Immunofluorescence analysis of HeLa cells stably expressing YFP–LAP2β, transfected with the indicated siRNA then stained with anti-mAb414 and DAPI (n = 3). Scale bars, 10 µm. d, Mask used to quantify nuclear pore formation by image-based flowcytometry (Imagestream). e, Imagestream analysis of HeLa cells transfected with the indicated siRNA, then stained with anti-mAb414 and DAPI. Nuclear pore intensity quantified by mask described in d. Representative images from two independent experiments, histogram and population averages displayed, graphical quantification of NPC intensity from the indicated number of gated cells (control, 3,045; CHMP2A-1, 1,256; CHMP2A-2, 2,152; CHMP2B, 5,237; UFD1-1, 4,146; UFD1-3, 4,325). Data are mean ± s.d.
Extended Data Figure 9 Requirements for nucleo-cytoplasmic compartmentalization.
a, Quantification of NE sealing from siRNA-treated cells as in Fig. 4b (control, 140 cells from 7 independent experiments; UFD1-1, 60 cells from 3 independent experiments, P = 0.044; UFD1-3, 60 cells from 3 independent experiments, P = 0.021; CHMP2B 40 cells from 2 independent experiments; two-tailed Student's t-test was used to assess significance at the 85-min time point). b, Resolved cell lysates from a were analysed by western blotting with the indicated antisera. c, NE integrity assay as performed with cells stably expressing mCh–H2B and GFP–NLS and transfected with the indicated siRNA. Differences in nucleo-cytoplasmic partitioning was assessed after plateau at the 65-min time point using a two-tailed Student's t-test (control, 79 cells from 4 independent experiments, CHMP2A-1, 60 cells from 3 independent experiments, P = 0.048; CHMP2A-2, 52 cells from 3 independent experiments, P = 0.011; CHMP3, 28 cells from 3 independent experiments, P = 0.028). d, e, HeLa cells stably expressing mCh–H2B and GFP–NLS were transfected with the indicated siRNA and imaged live. 60 min after anaphase onset, cytoplasmic signal was photo-ablated (T = 0) and recovery of cytoplasmic signal from the nuclear pool was calculated for the indicated conditions (cytoplasmic:nuclear ratio of GFP–NLS was normalized to T = 0; control, 21 cells from 4 independent experiments; CHMP2A-1, 24 cells from 4 independent experiments, P = 0.04; CHMP2A-2, 23 cells from 4 independent experiments, P = 0.05; CHMP3, 15 cells from 3 independent experiments, P = 0.004, two-tailed Student's t-test was used to assess significance after 10 min). Scale bars, 10 µm. f, Scoring of multinucleate and midbody-connected HeLa cells transfected with the indicated siRNA and stained with anti-tubulin and DAPI (300 cells analysed per condition, n = 4). Data are mean ± s.e.m. (a, c, d) and mean ± s.d. (f).
Extended Data Figure 10 Effect of CHMP2A depletion on NE discontinuities.
a, Presentation of reconstructed tomograms from Fig. 4d. b, CHMP2A-depleted cells exhibited more non-NPC discontinuities per unit area, while the number of NPC per unit area was constant. Tomograms as described in Fig. 4d, e were scored for discontinuities. The internal diameter of NPCs was slightly reduced in CHMP2A-depleted cells (control, 84 ± 7.6 nm, CHMP2A-1, 74 ± 8.8 nm; CHMP2A-2, 74 ± 5.7 nm). c, Schematic depicting topological equivalent of ESCRT-III-dependent membrane fusion events.
Supplementary information
CHMP2A forms a reticular network around telophase nuclei
Deconvolved 3D reconstruction of HeLa cells stained with anti-CHMP2A and DAPI and analysed by widefield microscopy, from Extended Data Figure 1C. (AVI 2486 kb)
GFP-CHMP4B transiently localises to telophase nuclei
Movie of GFP-CHMP4B localisation during the anaphase to telophase transition, from Extended Data Figure 1D. (AVI 1460 kb)
CHMP2A decorates nucleo-cytoplasmic channels
3D reconstruction of HeLa cells stained with anti-CHMP2A and DAPI and analysed by correlative light and electron tomography, From Figure 1D. (MOV 16147 kb)
Tomographic reconstruction of nascent nuclear envelope
3D reconstruction of HeLa cells stained with anti-CHMP2A and DAPI and analysed by correlative light and electron tomography, as depicted in Figure 1D and Extended Data Figure 2. (MOV 15633 kb)
Rights and permissions
About this article
Cite this article
Olmos, Y., Hodgson, L., Mantell, J. et al. ESCRT-III controls nuclear envelope reformation. Nature 522, 236–239 (2015). https://doi.org/10.1038/nature14503
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14503
This article is cited by
-
The plant unique ESCRT component FREE1 regulates autophagosome closure
Nature Communications (2023)
-
An ESCRT grommet cooperates with a diffusion barrier to maintain nuclear integrity
Nature Cell Biology (2023)
-
The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle
Cell Death & Differentiation (2022)
-
The ESCRT-III isoforms CHMP2A and CHMP2B display different effects on membranes upon polymerization
BMC Biology (2021)
-
PTEN alleviates maladaptive repair of renal tubular epithelial cells by restoring CHMP2A-mediated phagosome closure
Cell Death & Disease (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.