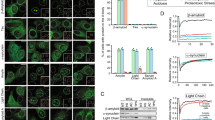
Abstract
Retromer is a multiprotein complex that trafficks cargo out of endosomes. The neuronal retromer traffics the amyloid-precursor protein (APP) away from endosomes, a site where APP is cleaved into pathogenic fragments in Alzheimer's disease. Here we determined whether pharmacological chaperones can enhance retromer stability and function. First, we relied on the crystal structures of retromer proteins to help identify the 'weak link' of the complex and to complete an in silico screen of small molecules predicted to enhance retromer stability. Among the hits, an in vitro assay identified one molecule that stabilized retromer against thermal denaturation. Second, we turned to cultured hippocampal neurons, showing that this small molecule increases the levels of retromer proteins, shifts APP away from the endosome, and decreases the pathogenic processing of APP. These findings show that pharmacological chaperones can enhance the function of a multiprotein complex and may have potential therapeutic implications for neurodegenerative diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Seaman, M.N. The retromer complex—endosomal protein recycling and beyond. J. Cell Sci. 125, 4693–4702 (2012).
Cullen, P.J. & Korswagen, H.C. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat. Cell Biol. 14, 29–37 (2012).
Seaman, M.N. Recycle your receptors with retromer. Trends Cell Biol. 15, 68–75 (2005).
Burd, C. & Cullen, P.J. Retromer: a master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol. 6, a016774 (2014).
Small, S.A. et al. Model-guided microarray implicates the retromer complex in Alzheimer's disease. Ann. Neurol. 58, 909–919 (2005).
Luo, Y. et al. Mice deficient in BACE1, the Alzheimer's β-secretase, have normal phenotype and abolished β-amyloid generation. Nat. Neurosci. 4, 231–232 (2001).
Jiang, Y. et al. Alzheimer's-related endosome dysfunction in Down syndrome is Aβ-independent but requires APP and is reversed by BACE-1 inhibition. Proc. Natl. Acad. Sci. USA 107, 1630–1635 (2010).
Nikolaev, A., McLaughlin, T., O'Leary, D.D. & Tessier-Lavigne, M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457, 981–989 (2009).
Jonsson, T. et al. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature 488, 96–99 (2012).
Nishitomi, K. et al. BACE1 inhibition reduces endogenous Aβ and alters APP processing in wild-type mice. J. Neurochem. 99, 1555–1563 (2006).
Small, S.A. & Gandy, S. Sorting through the cell biology of Alzheimer's disease: intracellular pathways to pathogenesis. Neuron 52, 15–31 (2006).
Bhalla, A. et al. The location and trafficking routes of the neuronal retromer and its role in amyloid precursor protein transport. Neurobiol. Dis. 47, 126–134 (2012).
Vieira, S.I. et al. Retrieval of the Alzheimer's amyloid precursor protein from the endosome to the TGN is S655 phosphorylation state-dependent and retromer-mediated. Mol. Neurodegener. 5, 40 (2010).
Muhammad, A. et al. Retromer deficiency observed in Alzheimer's disease causes hippocampal dysfunction, neurodegeneration, and Aβ accumulation. Proc. Natl. Acad. Sci. USA 105, 7327–7332 (2008).
Lane, R.F. et al. Diabetes-associated SorCS1 regulates Alzheimer's amyloid-β metabolism: evidence for involvement of SorL1 and the retromer complex. J. Neurosci. 30, 13110–13115 (2010).
Dodson, S.E. et al. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J. Neuropathol. Exp. Neurol. 65, 866–872 (2006).
Andersen, O.M. et al. Neuronal sorting protein–related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. USA 102, 13461–13466 (2005).
Fjorback, A.W. et al. Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J. Neurosci. 32, 1467–1480 (2012).
Rogaeva, E. et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 39, 168–177 (2007).
Vardarajan, B.N. et al. Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol. Aging 33, 2231.e15–2231.e30 (2012).
Arighi, C.N., Hartnell, L.M., Aguilar, R.C., Haft, C.R. & Bonifacino, J.S. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165, 123–133 (2004).
Seaman, M.N. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165, 111–122 (2004).
Vergés, M. et al. The mammalian retromer regulates transcytosis of the polymeric immunoglobulin receptor. Nat. Cell Biol. 6, 763–769 (2004).
Norwood, S.J. et al. Assembly and solution structure of the core retromer protein complex. Traffic 12, 56–71 (2011).
MacLeod, D.A. et al. RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson's disease risk. Neuron 77, 425–439 (2013); erratum 77, 994 (2013).
Ringe, D. & Petsko, G.A. What are pharmacological chaperones and why are they interesting? J. Biol. 8, 80 (2009).
Lieberman, R.L. et al. Structure of acid β-glucosidase with pharmacological chaperone provides insight into Gaucher disease. Nat. Chem. Biol. 3, 101–107 (2007).
Hierro, A. et al. Functional architecture of the retromer cargo-recognition complex. Nature 449, 1063–1067 (2007).
Mattos, C. & Ringe, D. Locating and characterizing binding sites on proteins. Nat. Biotechnol. 14, 595–599 (1996).
Collins, B.M., Skinner, C.F., Watson, P.J., Seaman, M.N. & Owen, D.J. Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nat. Struct. Mol. Biol. 12, 594–602 (2005).
Swarbrick, J.D. et al. VPS29 is not an active metallo-phosphatase but is a rigid scaffold required for retromer interaction with accessory proteins. PLoS ONE 6, e20420 (2011).
Laurie, A.T. & Jackson, R.M. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics 21, 1908–1916 (2005).
Collins, B.M. et al. Structure of Vps26B and mapping of its interaction with the retromer protein complex. Traffic 9, 366–379 (2008).
Shi, H., Rojas, R., Bonifacino, J.S. & Hurley, J.H. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain. Nat. Struct. Mol. Biol. 13, 540–548 (2006).
Landon, M.R., Lancia, D.R. Jr., Yu, J., Thiel, S.C. & Vajda, S. Identification of hot spots within druggable binding regions by computational solvent mapping of proteins. J. Med. Chem. 50, 1231–1240 (2007).
Mucke, L. et al. High-level neuronal expression of Aβ 1–42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J. Neurosci. 20, 4050–4058 (2000).
Acknowledgements
We thank the US National Institute on Aging/US National Institutes of Health (NIH) grants AG025161 and AG08702, The Alzheimer's Association, Developmental Therapeutics Program of the National Cancer Institute, Medkoo Biosciences, The Fidelity Biosciences Research Initiative and give special thanks to S. Weninger for advice and encouragement. We also thank The McKnight Endowment for Neuroscience, the Ellison Medical Foundation, and the Gottlieb Family Foundation.
Author information
Authors and Affiliations
Contributions
V.J.M. identified the retromer stabilizing site and carried out the in silico screens and designed and performed the in vitro retromer chaperone characterization; D.E.B. designed, coordinated and performed the in vivo retromer chaperone characterization; S.S. assisted with the immunohistochemical staining and genetic recombination in neurons; C.V. assisted with neuronal cultures; M.R.A. assisted with the in vitro thermal studies; V.M.P. assisted with the APP fragment analysis; R.T.S. helped with Aβ measurements; and G.A.P., D.R. and S.A.S. conceived of and supervised the studies and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Table 1, Supplementary Figures 1–7 and MedKoo certificate. (PDF 1821 kb)
Rights and permissions
About this article
Cite this article
Mecozzi, V., Berman, D., Simoes, S. et al. Pharmacological chaperones stabilize retromer to limit APP processing. Nat Chem Biol 10, 443–449 (2014). https://doi.org/10.1038/nchembio.1508
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1508
This article is cited by
-
VPS35 and α-Synuclein fail to interact to modulate neurodegeneration in rodent models of Parkinson’s disease
Molecular Neurodegeneration (2023)
-
Potential use of iPSCs for disease modeling, drug screening, and cell-based therapy for Alzheimer’s disease
Cellular & Molecular Biology Letters (2023)
-
Multi-omic approach characterises the neuroprotective role of retromer in regulating lysosomal health
Nature Communications (2023)
-
The understudied links of the retromer complex to age-related pathways
GeroScience (2022)
-
Conditional GWAS analysis to identify disorder-specific SNPs for psychiatric disorders
Molecular Psychiatry (2021)