Abstract
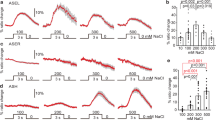
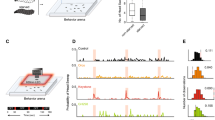
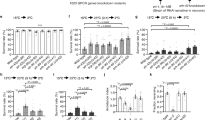
Dedicated neuronal circuits enable animals to engage in specific behavioral responses to environmental stimuli. We found that hypoxic stress enhanced gustatory sensory perception via previously unknown circuitry in Caenorhabditis elegans. The hypoxia-inducible transcription factor HIF-1 upregulated serotonin (5-HT) expression in specific sensory neurons that are not normally required for chemosensation. 5-HT subsequently promoted hypoxia-enhanced sensory perception by signaling through the metabotropic G protein–coupled receptor SER-7 in an unusual peripheral neuron, the M4 motor neuron. M4 relayed this information back into the CNS via the FMRFamide-related neuropeptide FLP-21 and its cognate receptor, NPR-1. Thus, physiological detection of hypoxia results in the activation of an additional, previously unrecognized circuit for processing sensory information that is not required for sensory processing under normoxic conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Joels, M. & Baram, T.Z. The neuro-symphony of stress. Nat. Rev. Neurosci. 10, 459–466 (2009).
de Kloet, E.R., Joels, M. & Holsboer, F. Stress and the brain: from adaptation to disease. Nat. Rev. Neurosci. 6, 463–475 (2005).
McEwen, B.S. Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol. Rev. 87, 873–904 (2007).
Chaouloff, F. Serotonin, stress and corticoids. J. Psychopharmacol. 14, 139–151 (2000).
Liang, B. et al. Serotonin targets the DAF-16/FOXO signaling pathway to modulate stress responses. Cell Metab. 4, 429–440 (2006).
Zhang, Y., Lu, H. & Bargmann, C.I. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184 (2005).
Chang, A.J. et al. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol. 4, e274 (2006).
Hukema, R.K., Rademakers, S. & Jansen, G. Gustatory plasticity in C. elegans involves integration of negative cues and NaCl taste mediated by serotonin, dopamine and glutamate. Learn. Mem. 15, 829–836 (2008).
Pocock, R. & Hobert, O. Oxygen levels affect axon guidance and neuronal migration in Caenorhabditis elegans. Nat. Neurosci. 11, 894–900 (2008).
Sze, J.Y. et al. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403, 560–564 (2000).
Semenza, G.L. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol. Med. 7, 345–350 (2001).
Epstein, A.C. et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 (2001).
Bishop, T. et al. Genetic analysis of pathways regulated by the von Hippel-Lindau tumor suppressor in Caenorhabditis elegans. PLoS Biol. 2, e289 (2004).
Shen, C. et al. Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J. Biol. Chem. 280, 20580–20588 (2005).
Semenza, G.L. et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537 (1996).
Rahman, M.S. & Thomas, P. Molecular cloning, characterization and expression of two tryptophan hydroxylase (TPH-1 and TPH-2) genes in the hypothalamus of Atlantic croaker: down-regulation after chronic exposure to hypoxia. Neuroscience 158, 751–765 (2009).
Poncet, L. et al. Activity of tryptophan hydroxylase and content of indolamines in discrete brain regions after a long-term hypoxic exposure in the rat. Brain Res. 765, 122–128 (1997).
Bargmann, C.I. & Horvitz, H.R. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7, 729–742 (1991).
Uchida, O. et al. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development 130, 1215–1224 (2003).
Hobson, R.J. et al. SER-7, a Caenorhabditis elegans 5-HT7-like receptor, is essential for the 5-HT stimulation of pharyngeal pumping and egg laying. Genetics 172, 159–169 (2006).
Hapiak, V.M. et al. Dual excitatory and inhibitory serotonergic inputs modulate egg laying in Caenorhabditis elegans. Genetics 181, 153–163 (2009).
Hobson, R.J. et al. SER-7b, a constitutively active Gα-coupled 5-HT7–like receptor expressed in the Caenorhabditis elegans M4 pharyngeal motorneuron. J. Neurochem. 87, 22–29 (2003).
Ray, P., Schnabel, R. & Okkema, P.G. Behavioral and synaptic defects in C. elegans lacking the NK-2 homeobox gene ceh-28. Dev. Neurobiol. 68, 421–433 (2008).
Mehta, R. et al. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science 324, 1196–1198 (2009).
Avery, L. & Horvitz, H.R. Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron 3, 473–485 (1989).
Albertson, D.G. & Thomson, J.N. The pharynx of Caenorhabditis elegans. Phil. Trans. R. Soc. Lond. B 275, 299–325 (1976).
Kim, K. & Li, C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J. Comp. Neurol. 475, 540–550 (2004).
Rogers, C. et al. Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat. Neurosci. 6, 1178–1185 (2003).
Gloria-Soria, A. & Azevedo, R.B. npr-1 regulates foraging and dispersal strategies in Caenorhabditis elegans. Curr. Biol. 18, 1694–1699 (2008).
de Bono, M. & Bargmann, C.I. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94, 679–689 (1998).
Coates, J.C. & de Bono, M. Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature 419, 925–929 (2002).
Hukema, R.K. et al. Antagonistic sensory cues generate gustatory plasticity in Caenorhabditis elegans. EMBO J. 25, 312–325 (2006).
Cheung, B.H. et al. Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr. Biol. 15, 905–917 (2005).
Pierce-Shimomura, J.T., Morse, T.M. & Lockery, S.R. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J. Neurosci. 19, 9557–9569 (1999).
Chang, A.J. & Bargmann, C.I. Hypoxia and the HIF-1 transcriptional pathway reorganize a neuronal circuit for oxygen-dependent behavior in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 105, 7321–7326 (2008).
Poncet, L. et al. Alteration in central and peripheral substance P- and neuropeptide Y–like immunoreactivity after chronic hypoxia in the rat. Brain Res. 733, 64–72 (1996).
Sarafi-Reinach, T.R. et al. The lin-11 LIM homeobox gene specifies olfactory and chemosensory neuron fates in C. elegans. Development 128, 3269–3281 (2001).
Sagasti, A. et al. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 13, 1794–1806 (1999).
Aspöck, G., Ruvkun, G. & Burglin, T.R. The Caenorhabditis elegans ems class homeobox gene ceh-2 is required for M3 pharynx motoneuron function. Development 130, 3369–3378 (2003).
Macosko, E.Z. et al. A hub-and-spoke circuit drives pheromone attraction and social behavior in C. elegans. Nature 458, 1171–1175 (2009).
Bargmann, C.I. & Horvitz, H.R. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7, 729–742 (1991).
Sze, J.Y. et al. Food and metabolic signaling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403, 560–564 (2000).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71 (1974).
Acknowledgements
We thank A. Boyanov and G. Baison for technical assistance, Q. Chen for expert microinjection help, I. Greenwald, M. De Bono and members of the Hobert laboratory for comments on the manuscript, and the Caenorhabditis Genetics Center and C. Bargmann for providing strains. This work was supported by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
R.P. initiated this study and conducted all of the experiments. R.P. and O.H. designed and discussed the experiments and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 1634 kb)
Rights and permissions
About this article
Cite this article
Pocock, R., Hobert, O. Hypoxia activates a latent circuit for processing gustatory information in C. elegans. Nat Neurosci 13, 610–614 (2010). https://doi.org/10.1038/nn.2537
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2537
This article is cited by
-
Neuromedin U signaling regulates retrieval of learned salt avoidance in a C. elegans gustatory circuit
Nature Communications (2020)
-
Serotonergic neuron ADF modulates avoidance behaviors by inhibiting sensory neurons in C. elegans
Pflügers Archiv - European Journal of Physiology (2019)
-
Novel animal model defines genetic contributions for neuron-to-neuron transfer of α-synuclein
Scientific Reports (2017)
-
Immunohistochemical analysis of the anterior nervous system of the free-living nematode Plectus spp. (Nematoda, Plectidae)
Zoomorphology (2017)
-
Serotonin promotes exploitation in complex environments by accelerating decision-making
BMC Biology (2016)