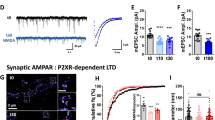
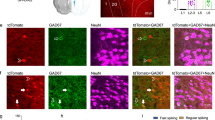
Abstract
NMDA receptors (NMDARs) are critical to synaptogenesis, neural circuitry and higher cognitive functions. A hallmark feature of NMDARs is an early postnatal developmental switch from those containing primarily GluN2B to primarily GluN2A subunits. Although the switch in phenotype has been an area of intense interest for two decades, the mechanisms that trigger it and the link between experience and the switch are unclear. Here we show a new role for the transcriptional repressor REST in the developmental switch of synaptic NMDARs. REST is activated at a critical window of time and acts via epigenetic remodeling to repress Grin2b expression and alter NMDAR properties at rat hippocampal synapses. Knockdown of REST in vivo prevented the decline in GluN2B and developmental switch in NMDARs. Maternal deprivation impaired REST activation and acquisition of the mature NMDAR phenotype. Thus, REST is essential for experience-dependent fine-tuning of genes involved in synaptic plasticity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cull-Candy, S.G. & Leszkiewicz, D.N. Role of distinct NMDA receptor subtypes at central synapses. Sci. STKE 2004, re16 (2004).
Paoletti, P. & Neyton, J. NMDA receptor subunits: function and pharmacology. Curr. Opin. Pharmacol. 7, 39–47 (2007).
Quinlan, E.M., Olstein, D.H. & Bear, M.F. Bidirectional, experience-dependent regulation of N-methyl-D-aspartate receptor subunit composition in the rat visual cortex during postnatal development. Proc. Natl. Acad. Sci. USA 96, 12876–12880 (1999).
Sheng, M., Cummings, J., Roldan, L.A., Jan, Y.N. & Jan, L.Y. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature 368, 144–147 (1994).
Williams, K., Russell, S.L., Shen, Y.M. & Molinoff, P.B. Developmental switch in the expression of NMDA receptors occurs in vivo and in vitro. Neuron 10, 267–278 (1993).
Yashiro, K. & Philpot, B.D. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology 55, 1081–1094 (2008).
Sobczyk, A., Scheuss, V. & Svoboda, K. NMDA receptor subunit-dependent [Ca2+] signaling in individual hippocampal dendritic spines. J. Neurosci. 25, 6037–6046 (2005).
Hall, B.J., Ripley, B. & Ghosh, A. NR2B signaling regulates the development of synaptic AMPA receptor current. J. Neurosci. 27, 13446–13456 (2007).
Gray, J.A. et al. Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron 71, 1085–1101 (2011).
Lee, M.C., Yasuda, R. & Ehlers, M.D. Metaplasticity at single glutamatergic synapses. Neuron 66, 859–870 (2010).
Xu, Z. et al. Metaplastic regulation of long-term potentiation/long-term depression threshold by activity-dependent changes of NR2A/NR2B ratio. J. Neurosci. 29, 8764–8773 (2009).
von Engelhardt, J. et al. Contribution of hippocampal and extra-hippocampal NR2B-containing NMDA receptors to performance on spatial learning tasks. Neuron 60, 846–860 (2008).
Espinosa, J.S., Wheeler, D.G., Tsien, R.W. & Luo, L. Uncoupling dendrite growth and patterning: single-cell knockout analysis of NMDA receptor 2B. Neuron 62, 205–217 (2009).
Barria, A. & Malinow, R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron 48, 289–301 (2005).
Borrelli, E., Nestler, E.J., Allis, C.D. & Sassone-Corsi, P. Decoding the epigenetic language of neuronal plasticity. Neuron 60, 961–974 (2008).
Levenson, J.M. & Sweatt, J.D. Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 6, 108–118 (2005).
Ballas, N., Grunseich, C., Lu, D.D., Speh, J.C. & Mandel, G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 (2005).
Ballas, N. & Mandel, G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr. Opin. Neurobiol. 15, 500–506 (2005).
Schoenherr, C.J. & Anderson, D.J. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267, 1360–1363 (1995).
Qiang, M., Rani, C.S. & Ticku, M.K. Neuron-restrictive silencer factor regulates the N-methyl-D-aspartate receptor 2B subunit gene in basal and ethanol-induced gene expression in fetal cortical neurons. Mol. Pharmacol. 67, 2115–2125 (2005).
Sasner, M. & Buonanno, A. Distinct N-methyl-D-aspartate receptor 2B subunit gene sequences confer neural and developmental specific expression. J. Biol. Chem. 271, 21316–21322 (1996).
Andres, M.E. et al. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 96, 9873–9878 (1999).
Bruce, A.W. et al. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. USA 101, 10458–10463 (2004).
Ooi, L. & Wood, I.C. Chromatin crosstalk in development and disease: lessons from REST. Nat. Rev. Genet. 8, 544–554 (2007).
Barria, A. & Malinow, R. Subunit-specific NMDA receptor trafficking to synapses. Neuron 35, 345–353 (2002).
Bellone, C. & Nicoll, R.A. Rapid bidirectional switching of synaptic NMDA receptors. Neuron 55, 779–785 (2007).
Matta, J.A., Ashby, M.C., Sanz-Clemente, A., Roche, K.W. & Isaac, J.T. mGluR5 and NMDA receptors drive the experience- and activity-dependent NMDA receptor NR2B to NR2A subunit switch. Neuron 70, 339–351 (2011).
Sanz-Clemente, A., Matta, J.A., Isaac, J.T. & Roche, K.W. Casein kinase 2 regulates the NR2 subunit composition of synaptic NMDA receptors. Neuron 67, 984–996 (2010).
Mierau, S.B., Meredith, R.M., Upton, A.L. & Paulsen, O. Dissociation of experience-dependent and -independent changes in excitatory synaptic transmission during development of barrel cortex. Proc. Natl. Acad. Sci. USA 101, 15518–15523 (2004).
Carmignoto, G. & Vicini, S. Activity-dependent decrease in NMDA receptor responses during development of the visual cortex. Science 258, 1007–1011 (1992).
Ku, H.Y., Huang, Y.F., Chao, P.H., Huang, C.C. & Hsu, K.S. Neonatal isolation delays the developmental decline of long-term depression in the CA1 region of rat hippocampus. Neuropsychopharmacology 33, 2847–2859 (2008).
Nase, G., Weishaupt, J., Stern, P., Singer, W. & Monyer, H. Genetic and epigenetic regulation of NMDA receptor expression in the rat visual cortex. Eur. J. Neurosci. 11, 4320–4326 (1999).
Philpot, B.D., Sekhar, A.K., Shouval, H.Z. & Bear, M.F. Visual experience and deprivation bidirectionally modify the composition and function of NMDA receptors in visual cortex. Neuron 29, 157–169 (2001).
Yu, H.B., Johnson, R., Kunarso, G. & Stanton, L.W. Coassembly of REST and its cofactors at sites of gene repression in embryonic stem cells. Genome Res. 21, 1284–1293 (2011).
Roopra, A., Qazi, R., Schoenike, B., Daley, T.J. & Morrison, J.F. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell 14, 727–738 (2004).
Bruce, A.W. et al. Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome Res. 19, 994–1005 (2009).
Bernstein, B.E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Noh, K.M. et al. Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc. Natl. Acad. Sci. USA 109, E962–E971 (2012).
Yeo, M., Berglund, K., Augustine, G. & Liedtke, W. Novel repression of Kcc2 transcription by REST-RE-1 controls developmental switch in neuronal chloride. J. Neurosci. 29, 14652–14662 (2009).
Monyer, H., Burnashev, N., Laurie, D.J., Sakmann, B. & Seeburg, P.H. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron 12, 529–540 (1994).
Liu, D., Diorio, J., Day, J.C., Francis, D.D. & Meaney, M.J. Maternal care, hippocampal synaptogenesis and cognitive development in rats. Nat. Neurosci. 3, 799–806 (2000).
Uchida, S. et al. Early life stress enhances behavioral vulnerability to stress through the activation of REST4-mediated gene transcription in the medial prefrontal cortex of rodents. J. Neurosci. 30, 15007–15018 (2010).
Desai, A., Turetsky, D., Vasudevan, K. & Buonanno, A. Analysis of transcriptional regulatory sequences of the N-methyl-D-aspartate receptor 2A subunit gene in cultured cortical neurons and transgenic mice. J. Biol. Chem. 277, 46374–46384 (2002).
Huang, Y., Myers, S.J. & Dingledine, R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 2, 867–872 (1999).
Prybylowski, K. et al. Relationship between availability of NMDA receptor subunits and their expression at the synapse. J. Neurosci. 22, 8902–8910 (2002).
Guardavaccaro, D. et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature 452, 365–369 (2008).
Westbrook, T.F. et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452, 370–374 (2008).
Huang, Z. et al. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat. Cell Biol. 13, 142–152 (2011).
Meaney, M.J. & Ferguson-Smith, A.C. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat. Neurosci. 13, 1313–1318 (2010).
Huot, R.L., Thrivikraman, K.V., Meaney, M.J. & Plotsky, P.M. Development of adult ethanol preference and anxiety as a consequence of neonatal maternal separation in Long Evans rats and reversal with antidepressant treatment. Psychopharmacology (Berl.) 158, 366–373 (2001).
Paoletti, P., Ascher, P. & Neyton, J. High-affinity zinc inhibition of NMDA NR1–NR2A receptors. J. Neurosci. 17, 5711–5725 (1997).
Pfaffl, M.W., Horgan, G.W. & Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36 (2002).
Acknowledgements
This work was supported by US National Institutes of Health grants to R.S.Z. (R01 NS46742 and NS20752) and P.E.C. (R01 MH081935) and support from the F.M. Kirby Foundation. A.E.C. was supported by a Ruth L Kirschstein Award from the US National Institute of Neurological Disorders and Stroke (F32NS071821). We thank K.-M. Noh (Albert Einstein College of Medicine) for providing the shRNA REST lentiviral construct, and A. Etgen and all the members of the Zukin and Castillo laboratories for their constructive comments on the data and the manuscript.
Author information
Authors and Affiliations
Contributions
A.R.-R. designed and performed all the biochemical experiments and analyzed the results. M.J.C. performed biochemical experiments. A.E.C. designed and performed all the electrophysiological experiments and analyzed the results. P.E.C. and R.S.Z. helped guide the research. A.R.-R., A.E.C., P.E.C. and R.S.Z. interpreted all the results and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Tables 1–11 (PDF 2066 kb)
Rights and permissions
About this article
Cite this article
Rodenas-Ruano, A., Chávez, A., Cossio, M. et al. REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci 15, 1382–1390 (2012). https://doi.org/10.1038/nn.3214
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3214
This article is cited by
-
REST in the Road Map of Brain Development
Cellular and Molecular Neurobiology (2023)
-
Neutral Sphingomyelinase 2 Mediates Oxidative Stress Effects on Astrocyte Senescence and Synaptic Plasticity Transcripts
Molecular Neurobiology (2022)
-
Neuroinflammation induces synaptic scaling through IL-1β-mediated activation of the transcriptional repressor REST/NRSF
Cell Death & Disease (2021)
-
Expression of G9a in Auditory Cortex Is Downregulated in a Rat Model of Age-Related Hearing Loss
Journal of Molecular Neuroscience (2021)
-
Neonatal Proinflammatory Stress and the Maturation of Intercellular Communication in the Hippocampus
Neuroscience and Behavioral Physiology (2020)