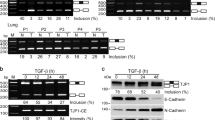
Abstract
The epithelial–mesenchymal transition (EMT), a prerequisite for cancer progression and metastasis formation, is regulated not only at the transcriptional but also at the post-transcriptional level, including at the level of alternative pre-mRNA splicing. Several recent studies have highlighted the involvement of splicing factors, including epithelial splicing regulatory proteins (Esrps) and RNA-binding Fox protein 2 (Rbfox2), in this process. Esrps regulate epithelial-specific splicing, and their expression is downregulated during EMT. By contrast, the role of Rbfox2 is controversial because Rbfox2 regulates epithelial as well as mesenchymal splicing events. Here, we have used several established cell culture models to investigate the functions of Rbfox2 during EMT. We demonstrate that induction of an EMT upregulates the expression of Rbfox2, which correlates with an increase in Rbfox2-regulated splicing events in the cortactin (Cttn), Pard3 and dynamin 2 (Dnm2) transcripts. At the same time, however, the epithelial-specific ability to splice the Enah, Slk and Tsc2 transcripts is either reduced or lost completely by Rbfox2, which might be due, in part, to downregulation of the expression of the Esrps cooperative factors. Depletion of Rbfox2 during EMT did not prevent the activation of transforming growth factor-β signaling, the upregulation of mesenchymal markers or changes in cell morphology toward a mesenchymal phenotype. In addition, this depletion did not influence cell migration. However, depletion of Rbfox2 in cells that have completed an EMT significantly reduced their invasive potential. Taken together, our results suggest that during an EMT, Rbfox2-regulated splicing shifts from epithelial-to mesenchymal-specific events, leading to a higher degree of tissue invasiveness.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Yang J, Weinberg RA . Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 2008; 14: 818–829.
Yilmaz M, Christofori G . EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev 2009; 28: 15–33.
Biamonti G, Bonomi S, Gallo S, Ghigna C . Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT). Cell Mol Life Sci 2012; 69: 2515–2526.
Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH . An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol 2009; 16: 130–137.
Huh GS, Hynes RO . Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev 1994; 8: 1561–1574.
Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL . Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol 2005; 25: 10005–10016.
Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev 2008; 22: 2550–2563.
Modafferi EF, Black DL . A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol 1997; 17: 6537–6545.
Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J 2003; 22: 905–912.
Nakahata S, Kawamoto S . Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res 2005; 33: 2078–2089.
Kuroyanagi H . Fox-1 family of RNA-binding proteins. Cell Mol Life Sci 2009; 66: 3895–3907.
Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol 2009; 16: 670–676.
Lapuk A, Marr H, Jakkula L, Pedro H, Bhattacharya S, Purdom E et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res 2010; 8: 961–974.
Ge X, Yamamoto S, Tsutsumi S, Midorikawa Y, Ihara S, Wang SM et al. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005; 86: 127–141.
Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 2011; 7: e1002218.
Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell 2005; 20: 881–890.
Valacca C, Bonomi S, Buratti E, Pedrotti S, Baralle FE, Sette C et al. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J Cell Biol 2010; 191: 87–99.
Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J 2010; 29: 3286–3300.
Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP . ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 2009; 33: 591–601.
Baraniak AP, Chen JR, Garcia-Blanco MA . Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol 2006; 26: 1209–1222.
Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME et al. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell 2001; 12: 27–36.
Miettinen PJ, Ebner R, Lopez AR, Derynck R . TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol 1994; 127 (6 Part 2): 2021–2036.
Lehembre F, Yilmaz M, Wicki A, Schomber T, Strittmatter K, Ziegler D et al. NCAM-induced focal adhesion assembly: a functional switch upon loss of E-cadherin. EMBO J 2008; 27: 2603–2615.
Waldmeier L, Meyer-Schaller N, Diepenbruck M, Christofori G . Py2T murine breast cancer cells, a versatile model of TGFbeta-induced EMT in vitro and in vivo. PLoS One 2012; 7: e48651.
Damianov A, Black DL . Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA 2010; 16: 405–416.
Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S et al. TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene 2012; 31: 3190–3201.
Kirkbride KC, Sung BH, Sinha S, Weaver AM . Cortactin: a multifunctional regulator of cellular invasiveness. Cell Adhes Migration 2011; 5: 187–198.
Hidalgo-Carcedo C, Hooper S, Chaudhry SI, Williamson P, Harrington K, Leitinger B et al. Collective cell migration requires suppression of actomyosin at cell-cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat Cell Biol 2011; 13: 49–58.
Venables JP . Unbalanced alternative splicing and its significance in cancer. Bioessays 2006; 28: 378–386.
Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest 2011; 121: 1064–1074.
Cheng C, Sharp PA . Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol 2006; 26: 362–370.
Moran-Jones K, Grindlay J, Jones M, Smith R, Norman JC . hnRNP A2 regulates alternative mRNA splicing of TP53INP2 to control invasive cell migration. Cancer Res 2009; 69: 9219–9227.
Goswami S, Philippar U, Sun D, Patsialou A, Avraham J, Wang W et al. Identification of invasion specific splice variants of the cytoskeletal protein Mena present in mammary tumor cells during invasion in vivo. Clin Exp Metastasis 2009; 26: 153–159.
Winter J, Lehmann T, Krauss S, Trockenbacher A, Kijas Z, Foerster J et al. Regulation of the MID1 protein function is fine-tuned by a complex pattern of alternative splicing. Hum Genet 2004; 114: 541–552.
Kruchten AE, McNiven MA . Dynamin as a mover and pincher during cell migration and invasion. J Cell Sci 2006; 119 (Part 9): 1683–1690.
Eppinga RD, Krueger EW, Weller SG, Zhang L, Cao H, McNiven MA . Increased expression of the large GTPase dynamin 2 potentiates metastatic migration and invasion of pancreatic ductal carcinoma. Oncogene 2012; 31: 1228–1241.
Feng H, Liu KW, Guo P, Zhang P, Cheng T, McNiven MA et al. Dynamin 2 mediates PDGFRalpha-SHP-2-promoted glioblastoma growth and invasion. Oncogene 2012; 31: 2691–2702.
Oh S, Shin S, Janknecht R . ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta 2012; 1826: 1–12.
Dikshit B, Irshad K, Madan E, Aggarwal N, Sarkar C, Chandra PS et al. FAT1 acts as an upstream regulator of oncogenic and inflammatory pathways, via PDCD4, in glioma cells. Oncogene 2013; 32: 3798–3808.
Nishikawa Y, Miyazaki T, Nakashiro K, Yamagata H, Isokane M, Goda H et al. Human FAT1 cadherin controls cell migration and invasion of oral squamous cell carcinoma through the localization of beta-catenin. Oncol Rep 2011; 26: 587–592.
Di Marcotullio L, Greco A, Mazza D, Canettieri G, Pietrosanti L, Infante P et al. Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene 2011; 30: 65–76.
Philippar U, Roussos ET, Oser M, Yamaguchi H, Kim HD, Giampieri S et al. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev Cell 2008; 15: 813–828.
Lemasters KE, Blech-Hermoni Y, Stillwagon SJ, Vajda NA, Ladd AN . Loss of muscleblind-like 1 promotes invasive mesenchyme formation in endocardial cushions by stimulating autocrine TGFbeta3. BMC Dev Biol 2012; 12: 22.
Dow LE, Elsum IA, King CL, Kinross KM, Richardson HE, Humbert PO . Loss of human Scribble cooperates with H-Ras to promote cell invasion through deregulation of MAPK signalling. Oncogene 2008; 27: 5988–6001.
Massimi P, Zori P, Roberts S, Banks L . Differential regulation of cell-cell contact, invasion and anoikis by hScrib and hDlg in keratinocytes. PLoS One 2012; 7: e40279.
Roovers K, Wagner S, Storbeck CJ, O’Reilly P, Lo V, Northey JJ et al. The Ste20-like kinase SLK is required for ErbB2-driven breast cancer cell motility. Oncogene 2009; 28: 2839–2848.
Barnes EA, Kenerson HL, Mak BC, Yeung RS . The loss of tuberin promotes cell invasion through the ss-catenin pathway. Am J Respir Cell Mol Biol 2010; 43: 617–627.
Liu H, Radisky DC, Nelson CM, Zhang H, Fata JE, Roth RA et al. Mechanism of Akt1 inhibition of breast cancer cell invasion reveals a protumorigenic role for TSC2. Proc Natl Acad Sci USA 2006; 103: 4134–4139.
Reed J, Stone M, Beadnell T, Ryu Y, Griffin T, Schwertfeger K . Fibroblast growth factor receptor 1 activation in mammary tumor cells promotes macrophage recruitment in a CXCL1-dependent manner. PLoS One 2012; 7: e45877.
Wesche J, Haglund K, Haugsten E . Fibroblast growth factors and their receptors in cancer. Biochem J 2011; 437: 199–213.
Guo M, Liu W, Serra S, Asa S, Ezzat S . FGFR2 isoforms support epithelial-stromal interactions in thyroid cancer progression. Cancer Res 2012; 72: 2017–2027.
Matsuda Y, Ueda J, Ishiwata T . Fibroblast growth factor receptor 2: expression, roles, and potential as a novel molecular target for colorectal cancer. Pathol Res Int 2012; 2012: 574768.
Jia YL, Shi L, Zhou JN, Fu CJ, Chen L, Yuan HF et al. Epimorphin promotes human hepatocellular carcinoma invasion and metastasis through activation of focal adhesion kinase/extracellular signal-regulated kinase/matrix metalloproteinase-9 axis. Hepatology 2011; 54: 1808–1818.
Hirata E, Yukinaga H, Kamioka Y, Arakawa Y, Miyamoto S, Okada T et al. In vivo fluorescence resonance energy transfer imaging reveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J Cell Sci 2012; 125 (Part 4): 858–868.
Acknowledgements
This work was supported by the Max-Planck Society. We are grateful to Rolf Kemler for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Braeutigam, C., Rago, L., Rolke, A. et al. The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene 33, 1082–1092 (2014). https://doi.org/10.1038/onc.2013.50
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.50
Keywords
This article is cited by
-
Role of epithelial splicing regulatory protein 1 in cancer progression
Cancer Cell International (2023)
-
Long noncoding RNA LINC01594 inhibits the CELF6-mediated splicing of oncogenic CD44 variants to promote colorectal cancer metastasis
Cell Death & Disease (2023)
-
RBFOX2 deregulation promotes pancreatic cancer progression and metastasis through alternative splicing
Nature Communications (2023)
-
Characterization of alternative mRNA splicing in cultured cell populations representing progressive stages of human fetal kidney development
Scientific Reports (2022)
-
Transcriptional and post-transcriptional control of epithelial-mesenchymal plasticity: why so many regulators?
Cellular and Molecular Life Sciences (2022)