Abstract
Aberrant Hedgehog/GLI signaling has been implicated in a diverse spectrum of human cancers, but its role in lung adenocarcinoma (LAC) is still under debate. We show that the downstream effector of the Hedgehog pathway, GLI1, is expressed in 76% of LACs, but in roughly half of these tumors, the canonical pathway activator, Smoothened, is expressed at low levels, possibly owing to epigenetic silencing. In LAC cells including the cancer stem cell compartment, we show that GLI1 is activated noncanonically by MAPK/ERK signaling. Different mechanisms can trigger the MAPK/ERK/GLI1 cascade including KRAS mutation and stimulation of NRP2 by VEGF produced by the cancer cells themselves in an autocrine loop or by stromal cells as paracrine cross talk. Suppression of GLI1, by silencing or drug-mediated, inhibits LAC cells proliferation, attenuates their stemness and increases their susceptibility to apoptosis in vitro and in vivo. These findings provide insight into the growth of LACs and point to GLI1 as a downstream effector for oncogenic pathways. Thus, strategies involving direct inhibition of GLI1 may be useful in the treatment of LACs.
Similar content being viewed by others
Introduction
The Hedgehog (HH)–GLI signaling is a conserved developmental pathway whose physiological roles in mammals consist of organogenesis, stem cell maintenance, and the upkeep and repair of adult tissues.1 Core pathway components include the secreted signaling proteins—Sonic HH (SHH), Indian HH or Desert HH—which bind to and inactivate the transmembrane receptor Patched (PTCH) on nearby target cells, annulling its repression of a second transmembrane receptor, Smoothened (SMO). The complex downstream signaling culminates in the expression of glioma-associated oncogene (GLI)-family transcription factors (GLI1, GLI2 and GLI3), whose targets comprise context-specific genes that regulate differentiation, proliferation and survival. GLI proteins also regulate the expression of genes encoding several HH pathway components, including GLI1 itself and PTCH1,2 thereby creating positive and negative feedback loops.
Aberrant activation of the HH–GLI cascade can be caused by a number of factors, including genetic alteration(s) involving individual pathway components (for example, mutations affecting PTCH or SMO,3 GLI1/2 amplification4, 5), and this anomalous signaling activity has been implicated in the initiation, progression and relapse of various types of cancer. In several epithelial forms of cancer, the tumor cells are reportedly dependent on autocrine HH–GLI stimulation.6 In other studies, however, HH-expressing neoplastic epithelial cells appear to be unresponsive to the ligand they produce, which acts instead on cells in the tumor stroma to create a microenvironment favorable for tumor growth.6 Inappropriate HH–GLI signaling also plays critical roles in maintaining the undifferentiated, self-renewing phenotype of the cancer stem cell (CSC) populations in hematological malignancies and solid tumors (including those of the lung, breast, pancreas, colon and brain).7, 8, 9, 10, 11 These tumor-initiating subpopulations are considered major drivers of tumor maintenance, metastasis, relapse and resistance to chemotherapy.12, 13 The HH–GLI pathway is thus an intriguing potential target for novel therapies for these cancers.
Several small-molecule inhibitors have been developed that suppress HH–GLI signaling via SMO blockade. Encouraging clinical results have been achieved in patients with medulloblastoma or basal cell carcinoma, which are associated with canonical HH–GLI signaling activation.14 In contrast, treatment of colorectal, gastric and ovarian cancers with these agents has not produced any significant responses.14 HH–GLI signaling is also known to play crucial cell-intrinsic roles in the development and maintenance of small-cell lung cancer.15 However, in patients with newly diagnosed extensive-stage small-cell lung cancer, the addition of SMO inhibition to standard chemotherapy regimens does not appear to offer any benefit in terms of progression-free or overall survival.16
Similar pictures are emerging of non-small-cell lung cancers (NSCLC), which account for approximately 85% of lung malignancies.17 Several lines of evidence suggest that HH–GLI signaling is a major player in NSCLC tumorigenesis and maintenance. The two main NSCLC histotypes, adenocarcinomas (LACs) and squamous cell carcinomas (LSCCs),18 express the GLI1 transcription activator19, 20, 21, 22 as well as the HH ligand SHH.23 In both histotypes, overexpression of HH–GLI pathway components has been linked to more aggressive behavior.21, 22, 24 Clinical trials are underway on the use of SMO inhibitors in advanced solid tumors including NSCLCs, but recently published preclinical data indicate that these agents are likely to exert few if any antitumoral effects on LSCC or LAC cells.24, 25 Importantly, however, LSCC cell lines that were unaffected by shRNA-mediated depletion of SMO or its pharmacological inhibition responded to genetic and pharmacological targeting of GLI with significant decreases in growth and markedly increased apoptosis.24 These findings are consistent with the noncanonical, SMO-independent activation of GLI-mediated transcription by input from other signaling pathways26, 27 such as the neuropilin-2/ERK (NRP2/ERK) cascade,28 K-Ras/ERK signaling29 and the mTOR/S6K1 pathway.30 These considerations induced us to take a closer look at the molecular mechanisms underlying HH–GLI pathway activation in LACs and the role played by this pathway in LAC CSCs.
Results
Expression of HH–GLI signaling pathway components in NSCLC
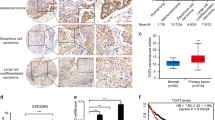
Interrogation of Oncomine31 microarray data sets on 1007 NSCLC tissues revealed positive correlation between GLI1 and SMO mRNA levels only in the LSCCs (Supplementary Figure S1a). Consistently, immunohistochemistry (IHC) analyses of human NSCLC tissue arrays disclosed GLI1 protein expression in most LACs (76%) and LSCCs (60%), but over half the GLI1-positive LACs exhibited weak SMO staining (Figures 1a and b). SHH was expressed in 28% of the LSCCs and 35% of the LACs (Supplementary Figure S1b).
HH–GLI pathway components in NCSLC. (a, b) IHC of SMO and GLI1 protein expression in human NSCLC tissue arrays (216 LACs, 291 LSCCs): (a) distribution of SMO/GLI1 phenotypes and (b) representative images of GLI1 and SMO staining. The LSCC sample in (b) is positive (H-score >50/300) for both proteins; the LAC displays only GLI1 positivity. Magnification × 20, inset × 40. Scale bar: 50 μm. (c, d) Western blots showing basal SMO and GLI1 protein expression in (c) commercial cell lines and (d) patient-derived CSCs from LSCC and LAC (see Supplementary Table S1 for cell genotypes). Bar graphs show densitometrically quantified band intensity values (n=3 or more) normalized to actin (loading control, LC). Asterisks show differences vs cell line with highest SMO expression. (e, f) Methylation levels in the proximal SMO regulatory region in (e) untreated LAC CSC lines and (f) commercial LAC cell lines before and after 5-AZA treatment. Cell lines are classified as SMOhigh (SMOH) or SMOlow (SMOL) based on findings shown in panels c and d. (g, h) Effect of 5-AZA-mediated demethylation on SMO expression (mRNA and protein) in (g) commercial LAC cell lines and (h) patient-derived LAC CSC lines. The mRNA level in each treated sample was calibrated against the corresponding basal level. LC: GAPDH. Bar graphs: densitometric analyses. *P<0.05; **P<0.01; ***P<0.001
GLI1 and SMO expression were then assessed in commercial cell lines and patient-derived CSC lines derived from NSCLCs (see Supplementary Table S1 for genotypes).32, 33 GLI1 was expressed in all cell lines. SMO expression varied and was particularly low in three LAC cell lines tested (H1437, CSC-3 and CSC-4—referred to hereafter as 'SMOlow cells'), as compared with both LSCC lines tested (H520 and CSC-1) and with the remaining three LAC lines ('SMOhigh cells') (Figures 1c and d).
Aberrant methylation of the SMO promoter region in LAC
Re-interrogation of the Oncomine31 data sets revealed no NSCLC-associated deletions/mutations involving the SMO region (chromosome 7, band 7q32.1) that might explain the heterogeneous expression documented in the LAC cell lines. To explore the possibility that the low expression in H1437, CSC-3 and CSC-4 reflected epigenetic silencing of SMO, which has been described in colorectal cancer,34, 35 we interrogated the MethHC database.36 Compared with patient-matched normal lung (NL) tissue, LACs (but not LSCCs) exhibited significantly higher SMO promoter methylation levels (Supplementary Figure S1c). Pyrosequencing was then used to quantify DNA methylation at 42 CpG dinucleotides in the SMO’s proximal regulatory region (Supplementary Figures S1d and e). Methylation levels in SMOlow LAC cells (~50% for CSC-4 and H1437) were significantly higher than those observed in SMOhigh cells (~5 for CSC-2, ~30% for H1734) (Figures 1e and f and Supplementary Figure S1e). Treatment with the demethylating agent 5-azacytidine (5-AZA) was followed by significantly reduced methylation levels and increased SMO expression (mRNA and protein) in SMOlow cells, but it had no effect on SMOhigh cells (Figures 1g and h).
Abrogation of SMO activity using shRNA-mediated knockdown (shSMO) or the SMO antagonist vismodegib37 reduced GLI1 transcription and cell proliferation in SMOhigh cells but had no effects in SMOlow cancer cells or in both SMOhigh and SMOlow CSCs (Supplementary Figures S2a–h). The ineffectiveness of SMO inhibition in LAC CSCs is consistent with previous reports.25
To investigate SMO function further, we treated LAC cancer cells with the SMO agonist purmorphamine.38, 39 As shown in Supplementary Figures S2i and j, purmorphamine produced increases in GLI1 expression and cell viability in SMOhigh cells but its effects in SMOlow cells were not significant. When cells were pretreated with 5-AZA, however, the effects of purmorphamine on GLI1 levels and cell viability were significant in the SMOlow cancer cells (Supplementary Figures S2i and j). These results indicate that SMO expression may be epigenetically silenced in LAC cells, that in LAC CSCs regardless of SMO levels, SMO inhibition is ineffective and that in LAC cancer cells SMO retained mild functionality.
GLI1 inhibition reduces LAC cell survival and CSC stemness features
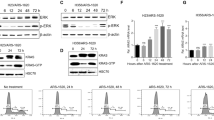
Direct targeting of GLI1 activity had more substantial effects in LAC cell lines and CSCs. Silencing of GLI1 significantly reduced viability in all lines tested (Figures 2a and b), and similar results were achieved when the cells were treated with GANT61, whose direct binding to GLI1 and GLI2 inhibits the latters’ DNA binding and their transcriptional output.40, 41 GANT61 is known to target GLI1 specifically,42 and numerous groups have demonstrated its high efficacy against human cancer cells (reviewed in Gonnissen et al.41). As shown in Figures 2c and d, GANT61 treatment significantly downregulated GLI1 levels in all LAC cell lines, regardless of their SMO expression status, and this inhibition was associated with significantly diminished cell viability and enhanced apoptosis (Supplementary Figure S3a).
GLI1 inhibition diminishes proliferation and stemness in LAC cells. (a–d) LAC cells viability is diminished by GLI1 knockdown in (a) commercial cell lines (siRNA-mediated) and (b) patient-derived CSCs (shRNA-mediated); similar reductions are seen in (c) cell lines and (d) CSCs treated for 48 h with the GLI1 antagonist GANT61 or DMSO (−). Bar graphs show viability (MTS assay); immunoblots show endogenous GLI1 protein levels. LC: GAPDH. Numbers below blots indicate densitometrically quantified protein expression. (e) Effects of GANT61 on oncosphere formation in LAC CSC lines before and after transduction with shGLI1. Bar graphs: frequencies of oncosphere-forming cells (OFC, % of seeded cells that formed oncospheres) under indicated experimental conditions are normalized to the frequency observed in shSCRAMBLE-infected cells treated with DMSO alone (controls, CTRL, expressed as 100%). Photomicrographs: Representative images of oncospheres observed in DMSO-treated CSCs infected with shGLI1 or shSCRAMBLE (CTRL). Scale bar: 50 μm. (f) Percentage of CSCs exhibiting high ALDH activity (ALDH+ cells) after GANT61 or CTRL treatment (AldeFluor assay). (g) Expression (mRNA and protein) of stem cell markers OCT4 and ABCG2 in GANT61- and CTRL-treated (dashed line) CSCs. LC: GAPDH. *P< 0.05. ALDH, aldehyde dehydrogenase.
Abrogation of GLI1 activity with shGLI1 or GANT61 treatment also diminished the self-renewal capacity of LAC CSCs, as reflected by their ability to form oncospheres (Figure 2e). Interestingly, when shGLI1-transduced CSCs were treated with GANT61, no additional decline was observed in the frequency of oncosphere-forming cells (OFCs), which suggests that the drug’s effects in LAC CSCs are mediated by its inhibition of GLI1. GANT61 treatment also significantly reduced the number of CSCs exhibiting high aldehyde dehydrogenase activity (Figure 2f), which is an established marker of lung CSCs.43, 44 The drug also reduced the expression of the lung CSC marker OCT4 (Figure 2g),32, 45, 46 and of the ATP binding cassette transporter ABCG2 (Figure 2g), which is commonly expressed by CSCs47, 48 and interestingly enough is reportedly regulated by HH pathway signaling.49, 50, 51
GLI1 inhibition reduces growth of LAC CSC-derived xenograft tumors, attenuates their stemness and induces apoptosis
Having established that in vitro GLI1 inhibition with GANT61 reduces the stemness and survival capacity of LAC CSCs, we investigated the drug’s in vivo effects on LAC CSC-derived xenograft tumors (XTs) (Supplementary Figure S4a). Tumors in GANT61-treated mice displayed significantly slower growth than their untreated counterparts (Figure 3a), together with decreased transcription of GLI1 and PTCH1 (Figure 3b), downregulated expression of OCT4 and ABCG2 (Figure 3c and Supplementary Figure S4b), attenuated cell proliferation (Figure 3d) and increased apoptosis (Figure 3e). As shown in Supplementary Figures S4c–e, the features of GANT61-treated XTs were replicated in XTs generated with LAC CSCs that had been stably infected with shGLI1. Compared with vector-infected controls, shGLI1 XTs exhibited slower growth (Supplementary Figure S4c), attenuated stemness (Supplementary Figure S4d) and decreased cell proliferation (Supplementary Figure S4e). These findings confirm that in vivo inhibition of GLI1 exerts antitumoral effects in LAC.
GLI1 inhibition produces antitumor effects in LAC CSC-derived XTs. (a) Up: growth of XTs generated in mice by s.c. flank injections of LAC CSC-2 (XT-2), CSC-3 (XT-3) and CSC-4 (XT-4). Results are shown for 4 weeks of treatment with i.p. GANT61 or vehicle (CTRL). Bottom: representative image of XTs after vehicle treatment of mice or after GANT61 treatment. Scale bar: 1 cm. (b) Human GLI1 and PTCH1 mRNA levels in XTs from GANT61-treated and CTRL (dashed line) mice. (c) GANT61’s effects on endogenous GLI1, ABCG2 and OCT4 protein expression in XTs. Bar graphs: densitometric analyses (see Supplementary Figure S3b for OCT4 IHC). (d, e) XT sections from GANT61-treated and CTRL mice were (d) immunostained for the proliferation marker KI-67 and (e) subjected to TUNEL assay for apoptosis. Representative results are shown (magnification: × 20; scale bar: 50 μm) with percentages of labeled cells (bar graphs on the right). Results are means±s.d. (n= 3). *P<0.05, **P<0.01 versus CTRLs.
Noncanonical GLI1 activation in LAC
The results of the experiments described thus far indicate that a substantial proportion of LACs are characterized by SMO-independent, GLI-mediated transcription, a phenomenon previously documented in other cancers. GLI1 regulation has been attributed to RAS/MEK signaling in breast cancer,28 melanoma52 and pancreatic ductal adenocarcinoma, and in the latter tumors, GLI1 proved to be indispensable for KRAS-induced carcinogenesis.53
Interestingly, in breast cancer, HH–GLI signaling was triggered in an autocrine manner by stimulation of the VEGF receptor neuropilin-2 (NRP2),28 whose expression in NSCLCs has been linked to unfavorable outcomes.54, 55, 56 In its role as a VEGF receptor, NRP2 specifically binds both VEGFA and VEGFC,57 and VEGF/NRP signaling impacts the biology of stem-like tumor cells in various types of cancer.58, 59, 60 We wondered whether oncogenic NRP2/ERK signaling might be the upstream cause of the noncanonical GLI1 activity in LAC cells.
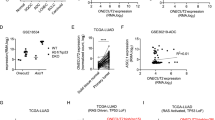
In support of this hypothesis, direct inhibition of MEK–ERK cascade in these cells with U012661 significantly reduced the levels of GLI1 and inhibited the phosphorylation of the MEK substrate ERK (Figures 4a and b). To determine whether the MEK–ERK axis directly regulates Gli1 activity through its modification, we investigated GLI1 phosphorylation in LAC cells treated with the selective MEK1/2 inhibitor U0126. As reported in Figure 4c, MEK inhibition caused a reduction of phosphorylated GLI1, corresponding to the active form of the transcription factor.30 Investigation of the subcellular localization of phosphorylated GLI1 in LAC cells showed positive nuclear staining for GLI1, which shifted to the cytoplasm after U0126 treatment (Figure 4d). To study whether ERK1 directly phosphorylates GLI1, we performed an in vitro kinase assay using the purified recombinant fragment containing the predicted ERK-sensing N-terminal region of GLI162 and ERK1. As shown in Figure 4e, the results revealed direct phosphorylation of GLI1, adding further support to the conclusion that MEK1–ERK1/2 activates GLI1 transcriptional activity in LAC cells.
MEK/ERK1/2 pathway regulates GLI1 phosphorylation. (a, b) Western blot analysis of endogenous levels of GLI1, phosphorylated ERK1/2 (p-ERK1/2) and actin (LC) in commercial LAC cell lines (H1734, H1437) and CSCs 2, 3, 4 after 48 h U0126 treatments. Bar graphs: densitometric analyses. Dashed line: CTRL treatment. *P<0.05; **P<0.01. (c) A549 and CSC-2 treated with U0126 for 4 h were lysed and subjected to (left) IP—western blot assay to assess endogenous levels of phosphorylated GLI1 (p-GLI1). Right: WB analysis of input of samples subjected to IP. (d) Immunofluorescence assay of endogenous GLI1 expression in A549 after 12 h of U0126 treatment. Nuclei are counterstained with Hoechst. Scale bars: 10 μm. (e) In vitro kinase assay. Recombinant GST-GLI1N (fragment 2-234) or GST were incubated with or without recombinant ERK1 for 20 min. Phosphorylation was revealed by PAN anti-pSer immunoblot. Total levels of GST and GST-GLI1N, revealed by α-GST immunoblot, are shown.
Experiments designed to elucidate the upstream role of NRP2 in the NRP2/ERK signaling showed that GLI1 expression was also significantly reduced in LAC cell lines after siRNA-mediated knockdown of NRP2 (Figure 5a). Similar decreases were seen in LAC CSCs following shRNA NRP2 knockdown (Figure 5b). Of note, NRP2 silencing was less effective in diminishing GLI1 expression in CSC-2, which harbor the oncogenic KRASG12C mutation (Supplementary Table S1 and Figures 5a and b).
Noncanonical activation of GLI1 in LAC cells. (a, b) Western blots of endogenous protein levels of NRP2, GLI1 and P-ERK1/2 and actin (LC) in commercial LAC cell lines (H1734, H1437) and CSCs 2, 3, 4 after siRNA-mediated (a) or short-hairpin-mediated (b) silencing of NRP2. Bar graphs: densitometric analyses. Dashed line: CTRL treatment. (c–f) Cell viability (MTS assay) in (c) LAC cell lines after siRNA silencing of NRP2, GLI1 and both; in (d) CSCs after short-hairpin mediated silencing of NRP2, GLI1 and both; and in (e) in LAC cell lines and (f) LAC CSCs treated for 48 h with GANT61, U0126 or both. *P<0.05, **P<0.01 vs sham-treated/sham-transfected controls.
As shown in Figure 5c, siNRP2 or siGLI1 in LAC cells both produced significant decreases in viability. In line with NRP2’s putative role as an upstream activator of GLI1 in these cells, we did not observe an additive effect when we combined NRP2 and GLI1 silencing.
To further clarify the role of KRAS mutation, we compared cell viability in CSC-2 (KRASG12C mut) and CSC-4 (KRASwt). GLI1 knockdown diminished the viability on both CSC lines, whereas shNRP2 decreased it only on CSC-4 (Figure 5d). No additive effects were achieved with combined shNRP2 and shGLI1 depletion.
U0126-mediated inhibition of the MAPK/ERK/GLI1 signaling also diminished the viability of LAC cell lines and CSCs. Similar effects were achieved with direct targeting of GLI1 with GANT61, and no additive effects were observed after their combination (Figures 5e and f).
Finally, as shown in Supplementary Figure S5a, the GLI1-modulating role of the NRP2/ERK axis was confirmed by the significant upregulation in cells stimulated with recombinant VEGF. Consistent with previous reports indicating that NRP2 and VEGFA may be direct targets of GLI1,28 transcript levels of both the receptor and its ligand in CSC-2 cells decreased significantly after siRNA-mediated GLI1 silencing (Supplementary Figure S5b).
Collectively, these findings support the existence of a self-amplifying autocrine signaling loop in LAC cells where VEGF/NRP2/MAPK/ERK signaling upregulates expression of GLI1 and GLI1 in turn enhances the expression and function of NRP2 and VEGF.
Our next set of experiments sought to document the existence of autocrine VEGF/NRP2/GLI1 signaling in vivo in LACs. Samples of LAC CSC-derived XTs were analyzed with quantitative PCR using assays specific for human transcripts to specifically characterize pathway component expression patterns in the cancer-cell compartments of tumors. Here we found NRP2 and VEGFA transcripts levels that significantly exceeded those found in normal human lung tissue (NL) (Figures 6a and b). VEGFA was previously shown to be often overexpressed in NSCLC.63 VEGFC was expressed at very low levels so we did not investigate its role further. Consistent with in vitro findings, XTs from GANT61-treated mice displayed significantly lower cancer-cell compartment levels of NRP2 and VEGFA than those from control animals (Supplementary Figure S6a).
Autocrine GLI1 activation and paracrine HH–GLI signaling in LAC. (a, b) Cancer-cell component of XTs: basal mRNA levels of (a) human NRP2 and (b) VEGFA vs levels in commercial normal human lung tissue (NL). Panel (a) shows IHC analysis of NRP2 expression in XT-3. Magnification: × 20, scale bar: 50 μm. (c) Human SHH mRNA levels in the cancer-cell component of XTs 2, 3 and 4 and normal human lung tissue (NL). (d) Representative IHC images of SHH and human nuclei (HuNu) staining in paraffin-embedded section of XT-3. Magnification: × 20, scale bar: 50 μm. (e) Relative expression of Gli1 and VegfA mRNA in normal murine lung fibroblasts (LF) treated for 24 h with recombinant murine Shh or BSA alone (CTRL). Results are reported as means±s.d. (n=4). (f, g) Stromal component of XTs 2–4 samples from GANT61-treated and CTRL mice: mRNA levels of murine (f) Gli1 and (g) VegfA. Gli1 levels are expressed vs normal murine LFs. Values in a–d and f–g are means±s.d. from four or more tumors. *P<0.05, **P<0.01, ***P>0.001.
The existence of an autocrine loop indicated by our in vitro findings was supported by our in vivo data (Figures 4 and 5).
Increasing evidence points to central roles for the tumor stroma in cancer maintenance.64 In particular, in situ hybridization studies have demonstrated the presence in LAC stroma of HH pathway components.19 In addition, exogenous SHH stimulation has been shown to activate canonical HH–GLI signaling in normal lung fibroblasts (LFs), upregulating their secretion of various oncogenic factors, including VEGF.23 In all the XTs we analyzed, SHH mRNA was highly expressed in the cancer-cell compartments of the tumors (Figures 6c and d), but it was undetectable in normal human lung (NL) and also in the XT stroma. As shown in Figure 6e, exposure of murine LFs to recombinant Shh markedly upregulated the transcription of Gli1 as well as that of VegfA. Analysis of the stromal compartment of the LAC XTs revealed transcription of Smo (at levels resembling those in normal murine lung and skin fibroblasts (LF, SF) (Supplementary Figure S6b), Gli1 (at levels significantly higher than those found in the normal fibroblasts) (Figure 6f) and VegfA (Figure 6g), and expression of the latter two genes was significantly downregulated in the stroma from GANT61-treated mice. These data suggest a paracrine SHH-mediated cross talk between the epithelial and stromal compartments of the tumor,27 which activates canonical HH–GLI signaling in the stroma, thereby upregulating the expression of VEGFA ligand available for interaction with epithelial-cell NRP2.
Collectively, the above findings point to the existence in LACs of a complex network in which GLI1 signaling can be activated by autocrine stimulation, genetic alterations (for example, activating KRAS mutations) and cross talk between the epithelial and stromal compartments of the tumor (Figure 7).
Noncanonical GLI1 signaling sustaining LAC growth and CSC maintenance. The HH pathway effector GLI1 can be activated even in the absence of SMO, the canonical upstream activator of HH signaling, whose expression is epigenetically silenced in many GLI1-expressing LACs. GLI1 serves as a downstream effector of oncogenic MAPK/ERK signaling, which can be triggered by ligand-binding events at NRP2 receptors expressed by the cancer cells. GLI1 then promotes features involved in tumor growth and CSC maintenance, including the transcription of known HH target genes (for example, ABCG2 and VEGFA). The VEGFA that binds epithelial-cell NRP2 receptors is secreted by the epithelial cells themselves, and this autocrine signaling loop is self-amplified by GLI1-upregulated transcription of both the receptor (NRP2) and its ligand (VEGFA). NRP2/ERK/GLI signaling can also be activated in a paracrine fashion by VEGFA secreted by cells in the tumor stroma. The stromal VEGFA production is the result of canonical SMO-dependent HH–GLI signaling triggered by SHH secreted by epithelial cells. The output of this stromal signaling includes VEGFA, which in turn bind tumor-cell NRP2, thereby triggering the noncanonical MAPK/ERK-mediated GLI1 activation illustrated in the upper panel.
Discussion
LAC, the world’s leading cause of cancer-related death, is characterized by high rates of somatic mutation and genomic rearrangement.65 The most common genetic alteration involves KRAS (33%). Aberrant signaling through several oncogenic pathways has also been described in LACs: that is, mitogen-activated protein kinases (MAPK) activation (76%); phosphatidylinositol 3-kinases (PI3K)–AKT–mammalian target of rapamycin (mTOR) activation (25%) and TP53 alteration (63%).65, 66
Aberrant HH–GLI signaling has been implicated in the maintenance of various epithelial cancers, including those of lung.14, 19, 39 In clinical trials, however, SMO inhibitors are producing disappointing results in several tumors.14, 16 Recently it has been shown that OFCs from LACs—unlike those from LSCCs—are unresponsive to drug-mediated SMO inhibition and authors concluded that these cells are probably not dependent on HH–GLI signaling.25 The LAC CSC we studied were also unaffected by SMO inhibition, but our data indicate that their refractoriness is not a reflection of HH–GLI signaling’s limited importance in these cells’ but rather of its noncanonical nature.
Our findings indicate that the canonical downstream effector of the HH pathway, GLI1, is expressed by approximately 75% of all LACs, but surprisingly, in roughly half of these GLI1-positive tumors, key upstream activator of this pathway, SMO, is expressed at low levels, at least in part owing to hypermethylation of its promoter. Even in SMOlow cells, however, we found evidence of GLI1-mediated transcription that was crucial for maintaining the growth and stemness features of LACs. Although virtually unaffected SMO targeting, this noncanonical HH–GLI signaling could be significantly impaired by GLI1 inhibition with antitumoral effects in vitro as well as in vivo.
As illustrated in Figure 7, our data show that noncanonical activation of GLI1 in LAC cells is at least partially the result of oncogenic signaling through the MAPK/ERK pathway, a phenomenon that has been described in other types of cancer.28, 29, 52, 53, 67 One of the mechanisms underlying activation of the MAPK/ERK/GLI1 cascade in LAC cells involves autocrine VEGFA stimulation of NRP2 receptors expressed by epithelial tumor cells. Autocrine VEGF/NRP2/GLI1 signaling has been implicated in the maintenance of tumor-initiating cells in breast cancer28 and esophageal adenocarcinoma.30 An autocrine loop involving NRP and VEGF has also been described in skin cancer, where the expression of VEGFA was shown to promote tumor growth not only by stimulating angiogenesis within the stroma but also by enhancing stemness features within the cancer-cell population.58
MAPK/ERK/GLI1 signaling can also be upregulated by paracrine stimulation of the epithelial-cell NRP2 receptors by VEGFA secreted by the stroma. This form of epithelial-cell/stromal-cell cross talk is mediated by the expression and secretion of SHH by the epithelial cells of LACs, which activates canonical HH–GLI signaling in stromal cells, upregulating the transcription of VEGFA and other GLI1 target genes.68, 69, 70 Our data build on and expands evidence of a potential role in lung cancer of HH–GLI signaling in the tumor stroma reported in earlier studies, where GLI1, SHH and PTCH transcripts were documented in LAC stromal cells.19 Our data also extend observations by Bermudez et al.23 that SHH secreted by lung cancer cells stimulates the proliferation of fibroblasts and their production of tumor-promoting factors, including VEGF.
Genetically determined, constitutive activation of KRAS is a well-known trigger of MAPK/ERK/GLI1 signaling.29 However, KRAS mutation per se is not necessarily associated with oncogenic KRAS activity.71 Functional comparison of various KRAS mutations in an in vivo system of vertebrate tumorigenesis showed tumorigenic effects for KRASG12C but not for KRASG13C.72 These results are in line with our finding that NRP2 silencing was less effective in reducing viability in LAC cells harboring the KRASG12C mutation than it was in KRASwt and KRASG13C cells.
KRAS mutation can also have important implications for paracrine stimulation of NRP2/GLI signaling in LAC epithelial cells. Mills et al.53 have shown that SHH secreted by pancreatic cancer cells activates canonical HH–GLI signaling in stromal cells, mediated in part by the upregulated expression of another GLI1 target gene. Interestingly, the expression of SHH by the cancer cells was found to be regulated by KRAS. This mechanism could parallel the role of stroma-produced VEGF as a target of SHH induced GLI1 activation in stromal cells in our experiments.
Our results point to GLI1 as an important factor for maintaining the biological aggressiveness of LACs, which is fully in line with the results of a recent meta-analysis documenting the negative prognostic repercussions of GLI1 expression in a wide range of solid tumor types.73 In our model, the tumor-promoting effects of GLI-mediated transcription are evident at multiple levels in LACs, including CSC and non-CSC components of the cancer-cell population and cells of the microenvironment that sustains them. Unlike approaches that target upstream regulators of GLI1 signaling, direct abrogation of GLI1 activity itself can exert antitumoral effects at all of these levels in LACs, in vitro and in vivo. GANT61 has been intensively used in various cancer research models; however, it has not been clinically tested.41 Other GLI antagonists, such as arsenic trioxide74 and atypical protein kinase C antagonists,75 have been used in clinical settings, but both require further investigation since they have molecular targets other than GLI1. A more recently identified drug that specifically inhibits GLI1, glabrescione B,76 is now undergoing preclinical investigation.
Prospective molecular profiling of LACs to assess their GLI1 status might be used to identify patients likely to benefit from GLI signaling blockade at different levels, with or without conventional chemotherapy or other promising biological approaches77, 78
Materials and methods
All commercial products were used according to suppliers’ instructions unless otherwise indicated.
Gene expression data set analyses
The Oncomine platform31 was used to interrogate data sets for LACs (Broet Lung, Lee Lung and Weir Lung) and LSCCs (TCGA Lung, Bass Lung, Kuner Lung, Larsen Lung and Lee Lung). Probes belonging to the same category were pooled (700 samples for LACs, 347 for LSCCs). Pearson’s r and correlation plots were generated to assess correlation between SMO and GLI1 expression. The TCGA Lung 2 data set was interrogated for SMO mutations in NSCLCs.
The MethHC database36 was interrogated to explore the methylation status of SMO CpG islands in NSCLC.
Cell cultures
Cells
All cell lines were subjected to quality-control testing. NSCLC cell lines A549, H1437 and H1734 were purchased from the ATCC (Milan, Italy); H520 were provided by Prof. M Broggini. Patient-derived CSC lines were obtained from primary LSCC (CSC-1) or LAC (CSC-2, -3 and -4). CSC-1–3 lines were characterized in Eramo et al.32 The CSC-4 line was isolated and characterized with internal review board approval at the University of Palermo. CSCs were maintained in advanced DMEM/F12 (Gibco Thermo Fisher Scientific, Waltham, MA, USA) supplemented with N2 (Gibco Thermo Fisher Scientific), B27 without vitamin A (Gibco Thermo Fisher Scientific), and 50 μg/ml insulin (Sigma-Aldrich, Milan, Italy), 20 ng/ml EGF and 10 ng/ml bFGF.
Murine lung and skin fibroblasts were isolated from NOD/SCID/IL2Rγnull mice as previously described.79
Treatments
Cells were treated (for 48 h unless otherwise specified) with the following: 10 μm 5-AZA (Sigma-Aldrich); 20 μm GANT61 (ENZO Lifesciences, Rome, Italy); 25 μm vismodegib (Selleckchem, Rome, Italy); 15 μm Purmorphamine (Selleckchem), 10 μm U0126 (TocrisBioscience, Milan, Italy); 100 ng/ml Recombinant VEFG (R&D Systems, Minneapolis, MN, USA); 25 μg/ml anti-VEGF (R&D Systems); and 3 μg/ml recombinant SHH (R&D Systems). SiRNA-mediated silencing was performed with smart pool on-target plus GLI1 (ID: L-003896) and NRP2 (L-003896) (Dharmacon, Milan, Italy); non-target siRNA was used as a control. For shRNA-mediated knockdown, PLKO lentiviral particles carrying shRNA (Sigma-Aldrich): MISSION shRNA-non-target control Transduction Particles (SCH002V) and three Lenti shGLI1: MISSION shRNA Clone TRCN0000020485, TRCN0000020486 and TRCN0000020487. Three lenti shSMO were used: MISSION shRNA Clone TRCN0000378375, TRCN0000378354, TRCN0000358090; Clones TRCN0000020487 (shGLI1) and TRCN0000358090 (shSMO) demonstrated the best knockdown efficiency with less off target effect.
Assays
To assess self-renewal, CSCs were plated at clonal density in 96-well plates. After 20 days the number of oncospheres was divided by the number of cells plated to determine the OFC frequency. Cell viability was assessed with MTS assay (Promega, Milan, Italy) and trypan blue exclusion. Aldehyde dehydrogenase activity was quantified with Aldefluor Kit (Stem Cell Technologies, Meda MB, Italy). All experiments were performed in at least three biological replicates.
Xenograft experiments
XTs were generated as previously described.32 CSCs (300 000 cells) were re-suspended in 100 μl of culture medium and Matrigel (1:1) (BD Pharmingen, Milan, Italy) and subcutaneously injected in the flanks of 6-week-old female NOD/SCID/IL2Rγnull mice (Charles River Laboratories, Calco, LC, Italy). Each experimental group consisted of six mice, allocated using a simple randomization method. Tumor volume (1/2(length × width2)) was assessed using digital caliper. Mice were treated with GANT61 (40 mg/kg, i.p.) re-suspended in ethanol and diluted to 100 μl per dose with 10% (2-hydroxypropyl)-β-cyclodextrin (vehicle) or vehicle alone (controls). Injections were administered twice a week for 4 weeks. All procedures were performed with the approval of the Ethics Committee for Animal Experimentation.
Immunohistochemistry
Standard-protocol IHC was done on Human Lung Cancer Survey tissue microarrays (US Biomax, Milan, Italy) and formalin-fixed paraffin-embedded (FFPE) XT tissues. The UltraTek HRP Anti-Polyvalent (DAB) Staining System (ScyTek, Logan, UT, USA) was used for blocking and detection; apoptosis was detected with TUNEL-POD kit (Roche, Monza, Italy). Labeling was detected with the UltraTek HRP Anti-Polyvalent (DAB) Staining System (ScyTek). Stained sections were examined with a Leica DM1000 microscope (Leica Microsystems Srl, Milan, Italy), photographed with a Jenoptik ProgRes SpeedXT Core 3 camera (Jenoptik AG, Vienna, Austria), and analyzed with ProgRes Capture Pro 2.8 software (Jenoptik AG).
Protein expression was evaluated with histoscore method (H-score): staining intensity was rated from 0 (negative) to 3 (strong) for each sample by two independent observers and the mean was used. H-score was calculated multiplying the number of cells within each category in at least three different × 200 fields. Human protein KI-67 expression and TUNEL scores were reported as percentages of labeled cells with respect to total cells.
Analysis of CpG island methylation status
DNA was extracted from LAC cells using Qiagen’s QIAamp DNA Mini Kit. Methylation at the CpG islands in the SMO promoter was assessed by pyrosequencing, performed according to standard procedures by EpigenDx Inc. (http://www.epigendx.com).
Western blotting
Western blots were performed according to standard procedures.80 Values are expressed as band integrals normalized to the strongest band and on the housekeeping gene.
Protein immunoprecipitation was performed as described in Canettieri et al.,81 incubating 1.5 mg of each total protein extract with anti-Gli1 and mouse IgG (Santa Cruz Biotechnology, Heidelberg, Germany), as a negative control. Images were acquired using the BioRad ChemiDoc MP Imaging System (BioRad, Hercules, CA, USA).
In vitro kinase assay
Recombinant GST-GLI1N (fragment 2-234 AA) and GST were produced as described previously.81 The in vitro kinase assay was performed as previously described.82 Recombinant ERK1 was purchased from Sigma-Aldrich (E7407).
mRNA analysis
mRNA levels were analyzed as described in Po et al.7 Endogenous controls were β-actin, GAPDH and HPRT for human samples, and Hprt1, β-2-microglobulin and GusB for murine. NL RNA was purchased from Thermo-Fisher Scientific (Waltham, MA, USA).
Immunofluorescence
Immunofluorescence was performed as described in Silvano et al.83 Images were acquired with a Carl Zeiss microscope (Axio Observer Z1) and AxioVision Digital Image Processing Software (Zeiss, Jena, Germany).
Antibodies
Santa Cruz Biotechnology: anti-SMO H-300 (sc-13943, IHC), anti-SMO N-19 (sc-6366, WB), anti-GLI1 H-300 (sc-20687, IHC), anti-actin I19 (sc-1616, WB), anti-NRP2 (sc-13117, IHC and WB), anti-p-ERK1/2 (sc-101761, WB) and anti-ABCG2 (sc-25821, WB). Abcam (Cambridge, UK): anti-SHH (ab53281, IHC), anti-GAPDH (ab8245, WB) anti-OCT4 (ab19857, WB and IHC) and Phospho—(Ser/Thr) (ab17464, WB-IP). Cell Signalling (Leiden, The Netherlands): anti-GLI1 (L42B10, WB). Thermo Fisher Scientific: Anti-KI-67 (RM-9106, IHC). EMD Millipore (Milan, Italy): Anti-human nuclei (MAB-1281, IHC) and anti-phospho-serine (#16-455 Kinase assay).
Statistical analysis
Results are expressed as means±s.d. from an appropriate number of experiments. Differences were analyzed by the Mann–Whitney U-test for non-parametric values with similar variance using the GraphPad Prism software Version 6.0 (La Jolla, CA, USA). Adjusted P-values of less than 0.05 were considered statistically significant.
References
Lee RTH, Zhao Z, Ingham PW . Hedgehog signalling. Development 2016; 143: 367–372.
Varjosalo M, Taipale J . Hedgehog: functions and mechanisms. Genes Dev 2008; 22: 2454–2472.
Katoh Y, Katoh M . Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med 2009; 9: 873–886.
Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, O'Brien SJ et al. Identification of an amplified, highly expressed gene in a human glioma. Science 1987; 236: 70–73.
Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC et al. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene 2005; 24: 4232–4242.
Theunissen J, de Sauvage F . Paracrine Hedgehog signaling in cancer. Cancer Res 2009; 69: 6007–6010.
Po A, Ferretti E, Miele E, De Smaele E, Paganelli A, Canettieri G et al. Hedgehog controls neural stem cells through p53-independent regulation of Nanog. EMBO J 2010; 29: 2646–2658.
Clement V, Sanchez P, De Tribolet N, Radovanovic I, Ruiz i Altaba A . HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol 2007; 17: 165–172.
Varnat F, Siegl‐Cachedenier I, Malerba M, Gervaz P, Ruiz i Altaba A . Loss of WNT‐TCF addiction and enhancement of HH‐GLI1 signalling define the metastatic transition of human colon carcinomas. EMBO Mol Med 2010; 2: 440–457.
Zbinden M, Duquet A, Lorente‐Trigos A, Ngwabyt SN, Borges I, Ruiz i Altaba A . NANOG regulates glioma stem cells and is essential in vivo acting in a cross‐functional network with GLI1 and p53. EMBO J 2010; 29: 2659–2674.
Justilien V, Fields A . Molecular pathways: novel approaches for improved therapeutic targeting of Hedgehog signaling in cancer stem cells. Clin Cancer Res 2015; 21: 505–513.
Garvalov B, Acker T . Cancer stem cells: a new framework for the design of tumor therapies. J Mol Med 2011; 89: 95–107.
Matchett K, Lappin T . Concise reviews: cancer stem cells: from concept to cure. Stem Cells 2014; 32: 2563–2570.
Amakye D, Jagani Z, Dorsch M . Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat Med 2013; 19: 1410–1422.
Park K, Martelotto L, Peifer M, Sos M, Karnezis A, Mahjoub M et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med 2011; 17: 1504–1508.
Belani CP, Dahlberg SE, Rudin CM, Fleisher M, Chen HX, Takebe N et al. Vismodegib or cixutumumab in combination with standard chemotherapy for patients with extensive‐stage small cell lung cancer: a trial of the ECOG‐ACRIN Cancer Research Group (E1508). Cancer 2016; 122: 2371–2378.
Davidson MR, Gazdar AF, Clarke BE . The pivotal role of pathology in the management of lung cancer. J Thorac Dis 2013; 5: S463–S478.
Johnson D . Setting the bar for therapeutic trials in non-small-cell lung cancer: how low can we go? J Clin Oncol 2014; 32: 1389–1391.
Singh S, Wang Z, Liang FD, Black K, Goetz J, Tokhunts R et al. Hedgehog-producing cancer cells respond to and require autocrine Hedgehog activity. Cancer Res 2011; 71: 4454–4463.
Raz G, Allen K, Kingsley C, Cherni I, Arora S, Watanabe A et al. Hedgehog signaling pathway molecules and ALDH1A1 expression in early-stage non-small cell lung cancer. Lung Cancer 2012; 76: 191–196.
Hwang J, Kang M, Yoo Y, Quan Y, Kim H, Oh S et al. The effects of sonic hedgehog signaling pathway components on non-small-cell lung cancer progression and clinical outcome. World J Surg Oncol 2014; 12: 268.
Yuan Z, Goetz J, Singh S, Ogden S, Petty W, Black C et al. Frequent requirement of hedgehog signaling in non-small cell lung carcinoma. Oncogene 2007; 26: 1046–1055.
Bermudez O, Hennen E, Koch I, Lindner M, Eickelberg O . Gli1 mediates lung cancer cell proliferation and Sonic Hedgehog-dependent mesenchymal cell activation. PLoS One 2013; 8: e63226.
Huang L, Walter V, Hayes DN, Onaitis M . Hedgehog–GLI signaling inhibition suppresses tumor growth in squamous lung cancer. Clin Cancer Res 2014; 20: 1566–1575.
Justilien V, Walsh M, Ali S, Thompson E, Murray N, Fields A . The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell 2014; 25: 139–151.
Lauth M, Toftgard R . Non-canonical activation of GLI transcription factors: implications for targeted anti-cancer therapy. Cell Cycle 2007; 6: 2458–2463.
Rubin L, de Sauvage F . Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov 2006; 5: 1026–1033.
Goel H, Pursell B, Chang C, Shaw L, Mao J, Simin K et al. GLI1 regulates a novel neuropilin-2/alpha6beta1 integrin based autocrine pathway that contributes to breast cancer initiation. EMBO Mol Med 2013; 5: 488–508.
Nolan-Stevaux O, Lau J, Truitt M, Chu G, Hebrok M, Fernandez-Zapico M et al. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev 2009; 23: 24–36.
Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell 2012; 21: 374–387.
Rhodes D, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs B et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18000 cancer gene expression profiles. Neoplasia 2007; 9: 166–180.
Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ 2008; 15: 504–514.
Sette G, Salvati V, Mottolese M, Visca P, Gallo E, Fecchi K et al. Tyr1068-phosphorylated epidermal growth factor receptor (EGFR) predicts cancer stem cell targeting by erlotinib in preclinical models of wild-type EGFR lung cancer. Cell Death Dis 2015; 6: 1850.
Zhu Y, James R, Peter A, Lomas C, Cheung F, Harrison D et al. Functional Smoothened is required for expression of GLI3 in colorectal carcinoma cells. Cancer Lett 2004; 207: 205–214.
Kober P, Bujko M, Olędzki J, Tysarowski A, Siedlecki J . Methyl‐CpG binding column‐based identification of nine genes hypermethylated in colorectal cancer. Mol Carcinog 2011; 50: 846–856.
Huang W-Y, Hsu S-D, Huang H-Y, Sun Y-M, Chou C-H, Weng S-L et al. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res 2014; 43: gku1151.
De Smaele E, Ferretti E, Gulino A . Vismodegib, a small-molecule inhibitor of the hedgehog pathway for the treatment of advanced cancers. Curr Opin Invest Drugs 2010; 11: 707–718.
Sinha S, Chen JK . Purmorphamine activates the Hedgehog pathway by targeting Smoothened. Nat Chem Biol 2006; 2: 29.
Rodriguez-Blanco J, Schilling NS, Tokhunts R, Giambelli C, Long J, Fei DL et al. The hedgehog processing pathway is required for NSCLC growth and survival. Oncogene 2013; 32: 2335–2345.
Lauth M, Bergström Å, Shimokawa T, Toftgård R . Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci USA 2007; 104: 8455–8460.
Gonnissen A, Isebaert S, Haustermans K . Targeting the hedgehog signaling pathway in cancer: beyond smoothened. Oncotarget 2015; 6: 13899.
Agyeman A, Jha B, Mazumdar T, Houghton J . Mode and specificity of binding of the small molecule GANT61 to GLI determines inhibition of GLI-DNA binding. Oncotarget 2014; 5: 4492.
Sullivan J, Spinola M, Dodge M, Raso M, Behrens C, Gao B et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res 2010; 70: 9937–9948.
Jiang F, Qiu Q, Khanna A, Todd N, Deepak J, Xing L et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res 2009; 7: 330–338.
Chen X, Fang F, Liou Y, Ng H . Zfp143 regulates Nanog through modulation of Oct4 binding. Stem Cells 2008; 26: 2759–2767.
Chiou S, Wang M, Chou Y, Chen C, Hong C, Hsieh W et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res 2010; 70: 10433–10444.
Fletcher J, Haber M, Henderson M, Norris M . ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer 2010; 10: 147–156.
Zinzi L, Contino M, Cantore M, Capparelli E, Leopoldo M, Colabufo N . ABC transporters in CSCs membranes as a novel target for treating tumor relapse. Front Pharmacol 2014; 5: 163.
Singh R, Kunkalla K, Qu C, Schlette E, Neelapu S, Samaniego F et al. ABCG2 is a direct transcriptional target of hedgehog signaling and involved in stroma-induced drug tolerance in diffuse large B-cell lymphoma. Oncogene 2011; 30: 4874–4886.
Ingram W, Crowther L, Little E, Freeman R, Harliwong I, Veleva D et al. ABC transporter activity linked to radiation resistance and molecular subtype in pediatric medulloblastoma. Exp Hematol Oncol 2013; 2: 26.
Chen Y, Bieber M, Teng N . Hedgehog signaling regulates drug sensitivity by targeting ABC transporters ABCB1 and ABCG2 in epithelial ovarian cancer. Mol Carcinog 2014; 53: 625–634.
Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V et al. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA 2007; 104: 5895–5900.
Mills L, Zhang Y, Marler R, Herreros-Villanueva M, Zhang L, Almada L et al. Loss of the transcription factor GLI1 identifies a signaling network in the tumor microenvironment mediating KRAS oncogene-induced transformation. J Biol Chem 2013; 288: 11786–11794.
Kawakami T, Tokunaga T, Hatanaka H, Kijima H, Yamazaki H, Abe Y et al. Neuropilin 1 and neuropilin 2 co‐expression is significantly correlated with increased vascularity and poor prognosis in nonsmall cell lung carcinoma. Cancer 2002; 95: 2196–2201.
Caunt M, Mak J, Liang W, Stawicki S, Pan Q, Tong R et al. Blocking neuropilin-2 function inhibits tumor cell metastasis. Cancer Cell 2008; 13: 331–342.
Nasarre P, Gemmill R, Potiron V, Roche J, Lu X, Barón A et al. Neuropilin-2 is upregulated in lung cancer cells during TGF-β1–induced epithelial–mesenchymal transition. Cancer Res 2013; 73: 7111–7121.
Favier B, Alam A, Barron P, Bonnin J, Laboudie P, Fons P et al. Neuropilin-2 interacts with VEGFR-2 and VEGFR-3 and promotes human endothelial cell survival and migration. Blood 2006; 108: 1243–1250.
Beck B, Driessens G, Goossens S, Youssef K, Kuchnio A, Caauwe A et al. A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumours. Nature 2011; 478: 399–403.
Glinka Y, Mohammed N, Subramaniam V, Jothy S, Prud’homme G . Neuropilin-1 is expressed by breast cancer stem-like cells and is linked to NF-κB activation and tumor sphere formation. Biochem Biophys Res Commun 2012; 425: 775–780.
Hamerlik P, Lathia J, Rasmussen R, Wu Q, Bartkova J, Lee M et al. Autocrine VEGF–VEGFR2–Neuropilin-1 signaling promotes glioma stem-like cell viability and tumor growth. J Exp Med 2012; 209: 507–520.
Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem 1998; 273: 18623–18632.
Riobo NA, Haines GM, Emerson CP . Protein kinase C-δ and mitogen-activated protein/extracellular signal–regulated kinase-1 control GLI activation in Hedgehog signaling. Cancer Res 2006; 66: 839–845.
Lantuéjoul S, Constantin B, Drabkin H, Brambilla C, Roche J, Brambilla E . Expression of VEGF, semaphorin SEMA3F, and their common receptors neuropilins NP1 and NP2 in preinvasive bronchial lesions, lung tumours, and cell lines. J Pathol 2003; 200: 336–347.
Kalluri R . The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016; 16: 582–598.
Network CGAR. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511: 543–550.
Ding L, Getz G, Wheeler D, Mardis E, McLellan M, Cibulskis K et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455: 1069–1075.
Seto M, Ohta M, Asaoka Y, Ikenoue T, Tada M, Miyabayashi K et al. Regulation of the hedgehog signaling by the mitogen‐activated protein kinase cascade in gastric cancer. Mol Carcinog 2009; 48: 703–712.
Cao X, Geradts J, Dewhirst M, Lo H . Upregulation of VEGF-A and CD24 gene expression by the tGLI1 transcription factor contributes to the aggressive behavior of breast cancer cells. Oncogene 2012; 31: 104–115.
Di Marcotullio L, Ferretti E, De Smaele E, Argenti B, Mincione C, Zazzeroni F et al. RENKCTD11 is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc Natl Acad Sci USA 2004; 101: 10833–10838.
Ingram W, Wicking C, Grimmond S, Forrest A, Wainwright B . Novel genes regulated by Sonic Hedgehog in pluripotent mesenchymal cells. Oncogene 2002; 21: 8196–8205.
Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N et al. A gene expression signature associated with 'K-Ras addiction' reveals regulators of EMT and tumor cell survival. Cancer Cell 2009; 15: 489–500.
Park JT, Johnson N, Liu S, Levesque M, Wang YJ, Ho H et al. Differential in vivo tumorigenicity of diverse KRAS mutations in vertebrate pancreas: a comprehensive survey. Oncogene 2015; 34: 2801–2806.
Cheng J, Gao J, Tao K, Yu P . Prognostic role of Gli1 expression in solid malignancies: a meta-analysis. Sci Rep 2016; 6: 22184.
Beauchamp E, Ringer L, Bulut G, Sajwan K, Hall M, Lee Y et al. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J Clin Invest 2011; 121: 148–160.
Atwood S, Li M, Lee A, Tang J, Oro A . GLI activation by atypical protein kinase C [igr]/[lgr] regulates the growth of basal cell carcinomas. Nature 2013; 494: 484–488.
Infante P, Mori M, Alfonsi R, Ghirga F, Aiello F, Toscano S et al. Gli1/DNA interaction is a druggable target for Hedgehog‐dependent tumors. EMBO J 2015; 34: 200–217.
Thomas A, Liu S, Subramaniam D, Giaccone G . Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol 2015; 12: 511–526.
Pinato DJ, Shiner RJ, White SD, Black JR, Trivedi P, Stebbing J et al. Intra-tumoral heterogeneity in the expression of programmed-death (PD) ligands in isogeneic primary and metastatic lung cancer: implications for immunotherapy. OncoImmunology 2016; 5: e1213934.
Seluanov A, Vaidya A, Gorbunova V . Establishing primary adult fibroblast cultures from rodents. J Vis Exp 2010; 44: e2033.
Ronci M, Catanzaro G, Pieroni L, Po A, Besharat ZM, Greco V et al. Proteomic analysis of human sonic hedgehog (SHH) medulloblastoma stem-like cells. Mol Biosyst 2015; 11: 1603–1611.
Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P et al. Histone deacetylase and Cullin3–RENKCTD11 ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol 2010; 12: 132–142.
D’Amico D, Antonucci L, Di Magno L, Coni S, Sdruscia G, Macone A et al. Non-canonical Hedgehog/AMPK-mediated control of polyamine metabolism supports neuronal and medulloblastoma cell growth. Dev Cell 2015; 35: 21–35.
Silvano M, Miele E, Valerio M, Casadei L, Begalli F, Campese A et al. Consequences of simulated microgravity in neural stem cells: biological effects and metabolic response. J Stem Cell Res Ther 2015; 5: 2.
Acknowledgements
We thank Alberto Gulino, friend, mentor and colleague, who passed away while the manuscript was in preparation, for his inspiring vision and continued support. This work was supported by Associazione Italiana Ricerca Cancro (AIRC and AIRC 5XMILLE), Ministry of University and Research (FIRB, PRIN and PON), Sapienza University of Rome (Ateneo), Pasteur Institute/Cenci Bolognetti Foundation, Umberto Di Mario ONLUS Foundation, Italian Institute of Technology (IIT) projects. Medical writing support was provided by ME Kent (EMWA) and funded by the Società Italiana Articoli Laboratorio S.r.l (Rome, Italy).
Author contributions
Conceptualization: EF, AP and CC; methodology: AP, MS and EM; investigation: AP, MS, EM, AE, VS, ZMB, MT, DC, GC, SC and LDiM; writing—original draft: AP, EF and EDeS; writing—review and editing: EF, IS and GC; funding acquisition: EF and EDeS and RDeM; resources: GS, MT and AE; supervision: EF.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Po, A., Silvano, M., Miele, E. et al. Noncanonical GLI1 signaling promotes stemness features and in vivo growth in lung adenocarcinoma. Oncogene 36, 4641–4652 (2017). https://doi.org/10.1038/onc.2017.91
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.91
This article is cited by
-
Gli1-mediated tumor cell-derived bFGF promotes tumor angiogenesis and pericyte coverage in non-small cell lung cancer
Journal of Experimental & Clinical Cancer Research (2024)
-
Hedgehog ligand and receptor cooperatively regulate EGFR stability and activity in non-small cell lung cancer
Cellular Oncology (2024)
-
N6-methyladenine-mediated aberrant activation of the lncRNA SOX2OT-GLI1 loop promotes non-small-cell lung cancer stemness
Cell Death Discovery (2023)
-
A clinicopathological exploration of Hedgehog signaling: implications in oral carcinogenesis
Journal of Cancer Research and Clinical Oncology (2023)
-
CPSF4 promotes tumor-initiating phenotype by enhancing VEGF/NRP2/TAZ signaling in lung cancer
Medical Oncology (2022)