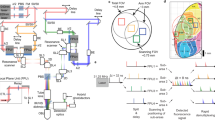
Abstract
Imaging neurons and neural circuits over large volumes at high speed and subcellular resolution is a difficult task. Incorporating a Bessel focus module into a two-photon fluorescence mesoscope, we achieved rapid volumetric imaging of neural activity over the mesoscale with synaptic resolution. We applied the technology to calcium imaging of entire dendritic spans of neurons as well as neural ensembles within multiple cortical regions over two hemispheres of the awake mouse brain.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The raw calcium imaging data for Figs. 2 and 3 are available at https://doi.org/10.6084/m9.figshare.11639787 and https://doi.org/10.6084/m9.figshare.11629098.v4.
Code availability
The custom CNMF-E processing codes in MATLAB are included as Supplementary Software.
References
Sofroniew, N. J., Flickinger, D., King, J. & Svoboda, K. A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. eLife 5, e14472 (2016).
Botcherby, E. J., Juškaitis, R. & Wilson, T. Scanning two photon fluorescence microscopy with extended depth of field. Opt. Commun. 268, 253–260 (2006).
Thériault, G., Cottet, M., Castonguay, A., McCarthy, N. & De Koninck, Y. Extended two-photon microscopy in live samples with Bessel beams: steadier focus, faster volume scans, and simpler stereoscopic imaging. Front. Cell. Neurosci. 8, 139 (2014).
Lu, R. et al. Video-rate volumetric functional imaging of the brain at synaptic resolution. Nat. Neurosci. 20, 620–628 (2017).
Lu, R., Tanimoto, M., Koyama, M. & Ji, N. 50 Hz volumetric functional imaging with continuously adjustable depth of focus. Biomed. Opt. Express 9, 1964–1976 (2018).
Welford, W. T. Use of annular apertures to increase focal depth. J. Opt. Soc. Am. 50, 749–752 (1960).
De Paola, V. et al. Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron 49, 861–875 (2006).
Chen, T.-W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Häusser, M., Spruston, N. & Stuart, G. J. Diversity and dynamics of dendritic signaling. Science 290, 739–744 (2000).
Manita, S., Miyakawa, H., Kitamura, K. & Murayama, M. Dendritic spikes in sensory perception. Front. Cell. Neurosci. 11, 29 (2017).
Reimer, J. et al. Pupil fluctuations track fast switching of cortical states during quiet wakefulness. Neuron 84, 355–362 (2014).
Pakan, J. M. et al. Behavioral-state modulation of inhibition is context-dependent and cell type specific in mouse visual cortex. eLife 5, e14985 (2016).
Pachitariu, M. et al. Suite2p: beyond 10,000 neurons with standard two-photon microscopy. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/061507 (2017).
Liu, H., Wu, Z., Li, X., Cai, D. & Huang, T. S. Constrained nonnegative matrix factorization for image representation. IEEE Trans. Pattern Anal. Mach. Intell. 34, 1299–1311 (2012).
Pnevmatikakis, E. A. et al. Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89, 285–299 (2016).
Zhou, P. et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife 7, e28728 (2018).
Ji, N., Freeman, J. & Smith, S. L. Technologies for imaging neural activity in large volumes. Nat. Neurosci. 19, 1154–1164 (2016).
Szalay, G. et al. Fast 3D imaging of spine, dendritic, and neuronal assemblies in behaving animals. Neuron 92, 723–738 (2016).
Stirman, J. N., Smith, I. T., Kudenov, M. W. & Smith, S. L. Wide field-of-view, multi-region, two-photon imaging of neuronal activity in the mammalian brain. Nat. Biotechnol. 34, 857–862 (2016).
Tsai, P. S. et al. Ultra-large field-of-view two-photon microscopy. Opt. Express 23, 13833–13847 (2015).
Horton, N. G. et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat. Photonics 7, 205–209 (2013).
Ouzounov, D. G. et al. In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nat. Methods 14, 388–390 (2017).
Rodríguez, C., Liang, Y., Lu, R. & Ji, N. Three-photon fluorescence microscopy with an axially elongated Bessel focus. Opt. Lett. 43, 1914–1917 (2018).
Thériault, G., De Koninck, Y. & McCarthy, N. Extended depth of field microscopy for rapid volumetric two-photon imaging. Opt. Express 21, 10095–10104 (2013).
Pnevmatikakis, E. A. & Giovannucci, A. NoRMCorre: an online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 291, 83–94 (2017).
Guizar-Sicairos, M., Thurman, S. T. & Fienup, J. R. Efficient subpixel image registration algorithms. Opt. Lett. 33, 156–158 (2008).
Kerlin, A. M., Andermann, M. L., Berezovskii, V. K. & Reid, R. C. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron 67, 858–871 (2010).
Acknowledgements
The authors thank N. Sofroniew for demonstrations on cranial window preparation; D. Flickinger for troubleshooting, optical alignments and helpful discussions; B. Mohar and A. Kerlin for help with system alignments and ScanImage operations; V. Custard for administrative support; J. Zheng for assistance with ScanImage; and M. Pachitariu for sharing the mesoscope and helpful discussions. R.L., Y.L., K.S. and N.J. were supported by the Howard Hughes Medical Institute. G.M. and N.J. were supported by NIH grant no. NINDS U01NS103489. P.Z. and L.P. were supported by NIH grant nos. R01-EB22913 and U01NS103489, the National Science Foundation (NeuroNex award no. DBI-1707398) and the Gatsby Foundation (GAT3708).
Author information
Authors and Affiliations
Contributions
N.J. conceived and supervised the project. R.L., Y.L., K.S. and N.J. designed the experiments. R.L. designed and built the Bessel module. Y.L. performed the mouse surgery. R.L. and Y.L. collected the data. P.Z. and L.P. developed the automatic ROI detection codes. R.L. and G.M. analyzed the data. All authors contributed to the preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The mesoscope (K.S.) and Bessel focus scanning (R.L. and N.J.) intellectual property has been licensed to Thorlabs by the Howard Hughes Medical Institute.
Additional information
Peer review information Nina Vogt was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Design and characterization of a two-photon mesoscope equipped with a Bessel module.
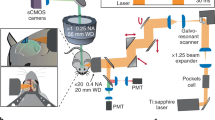
(a) Schematics of the mesoscope. BE: beam expander, BR: beam reducer, MTS: motorized translation stage to switch between Gaussian (yellow path) and Bessel (red path) modalities, PBS: polarizing beam splitter, QWP: quarter waveplate, RFO: remote focus objective, VC: voice coil, Res.: 12kHz resonant scanner, PR: pupil relay, DM: dichroic mirror, and IO: imaging objective. (b–e) Lateral and axial point spread functions of 0.6-NA Gaussian and 0.4-NA Bessel foci measured at different field positions with n = 9 0.2-µm-diameter beads. Error bar: s.e.m.; FWHM: full width at half maximum. (f, g) CAD model of the Bessel mesoscope and the Bessel beam module, respectively. See Supplementary Data for a complete part list and an Autodesk Inventor® model of the Bessel module.
Supplementary Figure 2 Adjustable Bessel focus.
Bessel focus with adjustable axial lengths by (a, b) displacing the lens L2 along its optical axis or (c, d) changing the beam diameter at the axicon. 2-µm-diameter beads were used for these measurements. All in vivo data were taken with a L2 displacement of 20 mm and a beam diameter of 4.5 mm.
Supplementary Figure 3 Detailed PSF measurements across the entire imaging field of view.
FWHMs of the lateral and axial PSFs of 0.6-NA Gaussian and 0.4-NA Bessel foci measured at different positions along (a-e) vertical, (f-j) horizontal, (k-o) and (p-t) diagonal directions, respectively, across the 5×5 mm2 field of view with 0.2-µm-diameter beads. N = 9 beads, error bar: s.e.m. The linearly polarized excitation light had its electric field along the y axis, leading to higher lateral resolution along the x axis. Data in Supplementary Fig. 1c are lateral FWHMs along horizontal direction with the excitation light polarized along x axis.
Supplementary Figure 4 Bessel focus scanning maintains synaptic resolution at depth.
(Left) Image stack of GCaMP6s+ dendrites and axons obtained with 3D Gaussian focus scanning, color-coded by depth; (Right) the same structures imaged with 2D Bessel scanning. Labels indicate depth below dura. Post-objective power: 120 mW for Bessel; 73 – 83.5 mW for Gaussian, exponentially increased with depth. Two experiments involving imaging of deep spines and boutons in one mouse were carried out, with similar results.
Supplementary Figure 5 Volumetric calcium imaging of a L2/3 neuron in a 396 µm × 400 µm × 200 µm volume.
(a) Maximum intensity projection of Gaussian image stack (101 images, 0.33-µm pixel, 2-µm z-step). (b) Composite of two Bessel images (0.33-µm pixel, 100-µm z step). (c) Calcium transients of example spines, dendrites, and soma. Red and blue arrows: local synaptic and dendritic transients, respectively. Blue lines: time points for local dendritic events. Insets: images of spines in (a), (b), and from a single Gaussian scan. (d) ΔF/F maps at three time points showing local dendritic and synaptic events (12 frame averages). Cyan arrows in (c,d): spines with activity coincident with dendritic branches D1, D2 or D3. Post-objective power: (a) 24-40 mW, increased exponentially with depth; (b) 120 mW.
Supplementary Figure 6 Detailed analysis of dendritic signals.
Same dataset as in Supplementary Fig. 5. (a) ΔF/F map, averaged across 12 consecutive frames (3.3 sec of image requisition). (b) Enlarged view of ΔF/F map in (a), with the ROIs for D1, D2 and D3 (used to calculate ΔF/F traces in Fig. 2g) shown in white outline. Magenta ROIs enclose dendritic spines. (c) Overlay of the ROIs on images of D1, D2 and D3 obtained with Bessel focus scanning. (d) Maximum intensity projection of Gaussian image stacks of the dendrites with structures color-coded by depth. Dendrite D1 spanned a depth range of z= 22 to 42 µm, D2 z= 8 to 80 µm, and D3 20 to 66 µm, respectively. (e, f) Fluorescence traces of segmented dendrites (D1, D2 and D3, black curves), spines (magenta curves), and dendritic shafts (green curves). The dendritic shaft signals were calculated by subtracting the magenta curves from the black curves.
Supplementary Figure 7 Simultaneous mesoscale volumetric functional imaging of distant neurites and synapses.
(a) Maximum intensity projection of a Gaussian image stack of neurites in an awake mouse cortex, with features color-coded by depth. FOVs P1 and P2 (white squares) were separated by ~ 2mm and selected for additional random-access functional imaging. (b) (Left panels) Maximum intensity projections of Gaussian image stacks at P1 and P2, with each volume extending over 400 µm × 425 µm × 200 µm and composed of 101 images with 0.5 µm pixel size and a z step of 2 µm. Features color-coded by depth. Volume rate: 0.1 Hz. (Right panels) Zoomed-in views of the structures within the four colored squares. Post-objective power: 36-48 mW, increased exponentially with depth. (c) Composite of 2 images acquired by scanning a Bessel focus at 0.5 µm per pixel and a z step size of 100 µm. Volume rate: 5.1 Hz. (Right panels) Zoomed-in views of the structures within the four colored squares. Post-objective power: 120 mW. (d) Raster plots of calcium transients detected from all spines and boutons in P1 and P2.
Supplementary Figure 8 Zoomed-in views of GCaMP6s+ GABAergic neurons imaged with Gaussian and Bessel modes.
Same data as in Fig. 3. (a) Gaussian and (b) Bessel images and (c, d) zoomed-in views of the neurons within the white squares. (e) Neurons identified by the CNMF-E method and color-coded by the Pearson’s correlation coefficients (CC) between their calcium traces and pupil size. n = 1 mouse was used for this experiment.
Supplementary Figure 9 Distributions of correlation coefficients between calcium transients and pupil area for neurons in different cortical areas and depths.
Histogram distributions of the Pearson’s correlation coefficient (CC) values of neuron in cortical areas C1-C5 and cortical depths D1-D6. Red and blue lines indicate means and medians of these distributions, respectively. D1-D6 had 1400, 1463, 1966, 2135, 1780 and 503 neurons, respectively; C1-C5 had 1460, 1972, 1866, 1812 and 2137 neurons, respectively. n = 1 mouse was used for this experiment.
Supplementary Figure 10 Analysis on GABAergic neuron ensemble activity within multiple cortical areas of an awake mouse using manually segmented ROIs.
(a, b) Same as Fig. 3a, b. (c) Calcium transients (ΔF/F) of 6,613 neurons in the six depth ranges, sorted by decreasing correlation with pupil area (top trace). (f) Map of the Pearson’s correlation coefficients (CC) for individual neurons, color-coded by its CC value. n = 1 mouse was used for this experiment.
Supplementary Figure 11 Distributions of Pearson’s correlation coefficients between calcium transients and pupil area for manually segmented neurons in different cortical areas and depths.
(a) Imaging areas. (b) Histograms of Pearson’s correlation coefficient (CC) values across brain areas and cortical depths, with their means and medians. (c) Summary of CC values of the all neuron-pupil pairs (N=6,613 pairs). (e) Mean of CCs of neurons in different cortical (top) areas and (bottom) depths. n = 1 mouse was used for this experiment.
Supplementary Figure 12 Mesoscale volumetric functional imaging of callosal-projection neurons across two hemispheres of an awake mouse.
(a) Schematic drawings illustrating the labeling of callosal-projection neurons and imaging FOVs in two hemispheres. (b) Maximum intensity projections (MIP) of images of two hemispheres and the cortical volumes P1, P2, and P3 at 50 – 250 µm below the dura, color-coded by depth. P1 and P2: two volumes symmetrically located across the midline, 400 µm × 400 µm × 200 µm and 400 µm × 800 µm × 200 µm, respectively; P3: a more posterior volume of 400 µm × 549 µm × 200 µm. (c) Images acquired by Gaussian and Bessel focus scanning of cortical volumes P1 – P3 (pixel size: 0.5 µm; z-step size: 2 µm for Gaussian). Gaussian images are MIPs with neurons color coded by depth. Volume rate: 0.08 Hz for Gaussian, 3.1 Hz for Bessel. Post-objective power: 36 mW for Gaussian, 108 mW for Bessel. (d) Calcium transients (ΔF/F) of all 189 neurons (48 in P1, 116 in P2, 25 in P3). (e) Traces of example neurons marked by arrows in (b). Red indicates neurons highly correlated with Neuron 1. CC: Pearson’s correlation coefficient. On population level, the cross-region activity correlation was higher between P1 and P2 than between P3 and P2 (Kolmogorov–Smirnov test, p = 0.0007, # of correlation coefficients: 5,568 for P1-P2, 2,900 for P3-P2; percentage of correlation coefficients above 0.2: 8% for P1-P2, 4% for P3-P2). Experiment carried out in n = 1 mouse.
Supplementary Figure 13 Comparison of neuropil subtraction methods.
ΔF/F of calcium transients of the same 9,247 neurons in Fig. 3 calculated after neuropil subtraction using (a) CNMF-E method and (b) the standard method of using signal from neighboring pixels as neuropil contribution. (c) Pearson’s correlation coefficient distributions of the calcium traces for neurons in different cortical depths range. Also see Supplementary Video 10 for side-by-side comparison of example depths (D1 and D2).
Supplementary information
Supplementary Information
Supplementary Figs. 1–13 and Notes 1–2
Supplementary Data
Supplementary CAD List; Part list and 3D models for Bessel beam module for the two-photon mesoscope.
Supplementary Software
Customized CNMF-E pipeline and the surrounding-pixel neuropil subtraction method (in MATLAB).
Supplementary Video 1
3D image stack of an infragranular neuron within a 301 µm × 450 µm × 612 µm volume, taken with serial 3D scanning of a Gaussian focus. Same data as in Fig. 2a. Volume rate: 0.06Hz; lateral pixel size: 0.4µm; axial step size: 2 µm.
Supplementary Video 2
Functional recording of an infragranular neuron in Fig. 2 using six 2D scanning of a Bessel focus. Structures are color-coded by depth as in Fig. 2b. Volume rate: 3.2 Hz; lateral pixel size: 0.4 µm; axial step size: 100 µm.
Supplementary Video 3
3D image stack of a layer 2/3 neuron within a 396 µm × 400 µm × 200 µm volume, taken with serial 3D scanning of a Gaussian focus. Same data as in Supplementary Fig. 5a. Volume rate: 0.06 Hz; lateral pixel size: 0.33 µm; axial step size: 2 µm.
Supplementary Video 4
Functional recording of the layer 2/3 neuron in Supplementary Fig. 5 using two 2D scanning of a Bessel focus. Structures are color-coded by depth as in Supplementary Fig. 5b. Volume rate: 3.2 Hz; lateral pixel size: 0.33 µm; axial step size: 100 µm.
Supplementary Video 5
Simultaneous mesoscale volumetric functional imaging of distant neurites and synapses. Inset during the first second of the video indicates the two regions P1 and P2, where neural activity was recorded. The rest of the video shows functional recording using Bessel focus scanning in P1 and P2, covering a depth of 0–200 µm below dura. Same data as in Supplementary Fig. 7.
Supplementary Video 6
3D image stacks of GABAergic neurons in a 3,020 µm × 1,500 µm × 600 µm volume, taken with serial 3D scanning of a Gaussian focus. Same data as in Fig. 3b. Volume rate: 0.02 Hz; lateral pixel size: 2 µm; axial step size: 2 µm.
Supplementary Video 7
Functional recording of calcium activity of GABAergic neurons within a 3,020 µm × 1,500 µm × 600 µm volume using six 2D scanning of a Bessel focus. Same data as in Fig. 3c. Volume rate: 1 Hz; lateral pixel size: 2 µm; axial step size: 100 µm.
Supplementary Video 8
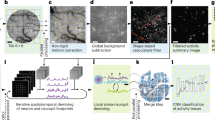
A customized CNMF-E pipeline that automatically segments ROIs and removes neuropil background. ‘Raw’ panel shows the raw data fed to the pipeline (zoomed in to visualize individual neurons enclosed by the orange box in Fig.3c). ‘Background’ (BG) frames model the baseline fluorescence and contaminating neuropil activities. Images in the remaining four panels were multiplied by a factor of two to improve visibility: ‘Denoised’ video displays the spatial and temporal features of neurons detected by the pipeline, and ‘Residual’ (Residual = Raw - BG - denoised) contains noise plus any sources not picked up by the pipeline; ‘Demixed’ video shows the same spatial/temporal information as ‘Denoised’, except that the extracted ROIs are pseudo color-coded to facilitate distinguishing nearby/overlapping neurons.
Supplementary Video 9
Spontaneous activity of callosal-projection neurons in three cortical regions across two hemispheres. Inset during the first second of the video indicates the three regions (P1, P2, and P3) across two hemispheres where neural activity was recorded. The rest of the video shows Bessel functional recording of neurons in these three regions covering a depth of 50–250 µm below dura. Left column: neurons 50–150 µm below dura; Right column: neurons 152–250 µm below dura. Same data as in Supplementary Fig. 12c-e.
Supplementary Video 10
Comparison of the CNMF-E and surrounding-pixel neuropil subtraction methods. The top panel shows ΔF/F traces with background automatically subtracted by CNMF-E and the bottom panel with surrounding-pixel neuropil subtraction. Traces of neurons in depths D1 and D2 were presented.
Rights and permissions
About this article
Cite this article
Lu, R., Liang, Y., Meng, G. et al. Rapid mesoscale volumetric imaging of neural activity with synaptic resolution. Nat Methods 17, 291–294 (2020). https://doi.org/10.1038/s41592-020-0760-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0760-9
This article is cited by
-
Large-scale recording of neuronal activity in freely-moving mice at cellular resolution
Nature Communications (2023)
-
Rapid detection of neurons in widefield calcium imaging datasets after training with synthetic data
Nature Methods (2023)
-
Multiregion neuronal activity: the forest and the trees
Nature Reviews Neuroscience (2022)
-
The neurons that restore walking after paralysis
Nature (2022)
-
Flexible simultaneous mesoscale two-photon imaging of neural activity at high speeds
Nature Communications (2021)