Abstract
Congenital profound deafness has a known genetic origin in more than 50% of all cases. The majority of the non syndromic hearing loss (NSHL) show an autosomal recessive inheritance. Mutations in the GJB2 gene (connexin 26) account for more than 50% of the recessive non syndromic deafness (DFNB1) among 30 loci. Other connexin genes have been more rarely involved and attention was given here to the GJB6 gene (connexin 30). We show that homozygous deletion of a minimal 150 kb region encompassing this gene causes NSHL. More strikingly, association of this deletion in trans of the GJB2 gene 35delG or E47X mutations is also associated with NSHL.
Similar content being viewed by others
Introduction
The majority of the non syndromic hearing loss (NSHL) show an autosomal recessive inheritance,1 and alterations in the GJB2 gene, encoding connexin 26, is recognised in half of the cases. Among 50 mutations identified so far, some are major common mutations found in specific populations.2,3,4,5 A subset of patients is reported to carry a single heterozygous mutation of this gene, possibly reflecting the carrier frequency, though digenic inheritance has also been suggested.6 More rarely, GJB2 has been involved in non syndromic deafness with an autosomal dominant pattern of inheritance (DFNA3).7 Connexin 26 is a member of a large family of gap junction channel proteins that allow intercellular communication and regulate the passage of ions and small metabolites. The connexins form hexameric hemichannels (called connexon) which have to dock with a connexon from an adjacent cell to form a functional channel.8 Three other gap junction genes GJB1, GJB3 and GJB6 respectively encoding connexins 32, 31 and 30, are known to cause deafness. One mutation in the GJB1 gene is suspected to be responsible for deafness associated with peripheral neuropathy.9 Mutations in GJB3 are responsible for dominant or recessive deafness.10,11 Finally, one mutation in GJB6 causes hearing loss by a dominant negative effect.12 As the two genes GJB6 and GJB2 are lying ∼35 kb apart, they both define the DFNA3 locus. Remarkably, GJB2, GJB3 and GJB6 are also involved in inherited skin disorders13,14,15 underlining the importance of these connexins in very distinct tissues.
Connexins 26 and 30 are structurally highly related.12,16 Moreover, expression analyses performed in mouse and rat show that Cx26 and Cx30, as well as Cx31 and Cx32, form two co-expression groups.17,18 Co-expression of Cx26 and Cx30 in the inner ear is thought to be a prerequisite for the maturation of the cochlea.18,19 Their co-expression is also detectable in human embryo cochlea after the 22nd week.19,20 The co-expression of these two connexins also favoured the hypothesis that their connexons may form both homo- and/or hetero-channels.21
Patients and methods
Patients
The five patients presented in this study were referred to us by the Genetic counselling and the Ear Nose and Throat Clinics. Clinical history of each individual was collected and audiograms were performed. For the five patients the hearing loss was sensorineural, bilateral, prelingual and profound. Informed consent was obtained from each individual.
PCR analysis
Genomic DNA was extracted from blood samples using the Nucleon BACC3 DNA extraction kit (Amersham Pharmacia Biotech, Piscataway, NJ, USA) according to the manufacturer's instructions.
Mutation analysis in the GJB2 gene was performed by PCR of the non coding exon and the coding exon, followed by DHPLC analysis (WAVE DNA Fragment Analysis System, Transgenomic) and sequencing (ABI 310 sequencer, PE biosystem).
The coding region of the GJB6 gene (CX30) was amplified in three overlapping fragments R1, R2 and R3 of 431, 410, 333 bp, respectively. Primer sequences are as follow: (CX30-R1: ctttgcccacttttgtctgt/gacccctctatccgaacctt; CX30-R2: cctccagctgatcttcgtct/gcagcaggtagcacaactct); CX30-R3: gccccaaccttgttgactgc/gttggtattgccttctggag. Primer sequences for D13S1189 analysis were retrieved from NCBI.
Quantification
We used Light Cycler Master SYBR Green I on a LightCycler system (Roche Diagnostics). Quantification analysis could not be performed for D13S175 which is a microsatellite marker. Primers used for CX26 gene analysis have been described.2 Primers used for CX30-R1/CX30-R3 analysis are indicated above. Primers used for DMD exon 60 are already published.22 A negative control without DNA sample and a standard curve (31.25 ng – 15.6 ng – 7.8 ng – 3.9 ng – 1.95 ng – 0.975 ng – 0.4875 ng) were included in every test. For each DNA sample, four replicate points were performed and expressed in mean±SD (standard deviation). In addition, quantification was performed twice for each sample to confirm the results.
Electronic-database information
The accession numbers and URLs for this article are as follows: http://www.iro.es/deafness/ (connexin-deafness homepage); http://www.uia.ac.be/dnalab/hhh (Hereditary Hearing Loss Homepage); DFNB1 {MIM 220290} DFNA3 {MIM 601544}; GJB1 {MIM 304040} GJB2 {MIM 121011} GJB3 {MIM 603324} GJB6 {MIM 604418}; Genomic sequence of the GJB2 and GJB6 region: http://www.ncbi.nlm.nih.gov/; Chromosome 13 map/contig view: http://www.ensembl.org/Homo_sapiens/contigview?chr=chr13;GenBank:http://www.ncbi.nlm.nih.gov/
Results
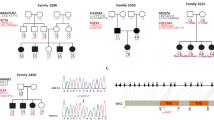
Because no mutation in the GJB2 gene could be found in patient SU1, a sporadic case presenting congenital profound deafness, we decided to study other deafness genes, in particular GJB6, because of its structural and expression similarities with GJB2. We then identified for the first time the deletion reported here. When PCR analysis of the GJB6 coding region was performed, only the 3′ end of the gene (CX30-R3) could be detected, whereas the 5′ end (CX30-R1 and CX30-R2) was not amplifying, thus suggesting a homozygous deletion in GJB6 (Table 1). We excluded uniparental disomy as a possible origin for homozygosity by microsatellite marker analyses (listed in Figure 1). Absence of amplification was then noticed for marker D13S175, lying 5′ to GJB6. Furthermore, this deletion appeared to extend beyond the non polymorphic marker D13S1189 (Table 1 and Figure 1) suggesting that the deleted region covers at least 150 kb, based on publicly available genomic sequence NT_009917. Discrepancies and ambiguities within the region adjacent to D13S1189 does not permit yet precise mapping of the other deletion border (Figure 1).
Schematic representation of the DFNB1 region and the deletion. The order of the markers is given according to the contig view (Ensembl project from the Sanger Institute). The distances between genes and markers from NT_009917 contig are indicated. CX26 (GJB2) and CX30 (GJB6) genes are indicated by arrows. The deletion is indicated: the centromeric breakpoint is within fragment CX30-R2 in the coding exon, the telomeric breakpoint is unknown but lies beyond marker D13S1189.
Independently, we studied patient SU5 who was referred for congenital profound deafness. GJB2 sequence analysis revealed the presence of the 35delG mutation in heterozygous state with no other mutation within the gene. Family study showed that, like her unaffected brother, she had inherited the GJB2 35delG mutation from her unaffected mother (Figure 2A). Haplotypes analysis was then performed in the DFNB1 region (encompassing the GJB2 gene) to exclude or confirm involvement of this region in deafness with the possible presence of a cryptic mutation (Figure 2A). Involvement of DFNB1 region could not be excluded as the proposita had inherited from her unaffected father a different chromosome than her brother. However, and surprisingly, the proposita SU5 showed a single peak for D13S175 defining allele 103 suggesting either a neomutation of the paternal inherited allele or absence of its amplification. The father had been first defined as homozygous 101/101.
Haplotypes analysis in the DFNB1 region. (A) SU5 family and (B) SU172 family. Carriers of the GJB2 35delG or E47X mutation are indicated by black fillings; carriers of the GJB6 deletion are indicated by diagonal hatching. Haplotypes carrying the deletion are boxed. The size of each polymorphic allele is indicated in base pair (bp). The proposita (SU5) and propositus (SU172) are indicated by a thick arrow. N: normal; D: deleted.
Putting these data together, we then wondered if D13S175 could be deleted along with the major part of GJB6 in SU5, as in SU1, but in heterozygous state. To address this hypothesis, we performed real time quantitative PCR using Light-Cycler (Roche diagnostics) on all the deleted PCR fragments observed in SU1 except D13S175 marker. The results, presented on Table 2, show that SU5 presents two copies of the 3′ end of the GJB6 gene (CX30-R3) but only one copy of the 5′ coding region of the GJB6 gene (CX30-R1) and D13S1189 and therefore concluded that only one allele was present for D13S175 (Tables 1 and 2). Moreover, quantitative PCR performed on the parents and the brother of the proposita showed that this deletion was inherited from the unaffected father (SU16) who was, therefore, hemizygote for D13S175 (Figure 2A and Table 2).
Three additional unrelated patients (SU47, SU145 and SU172) also referred for profound congenital deafness showing one heterozygous mutation in the GJB2 gene (35delG or E47X) were then tested for the D13S175 and revealed either hemizygosity or homozygosity for one allele (Table 1). Consequently, analysis of the region by quantitative real time PCR evidenced that they were also deleted in the same region including marker D13S1189 (Table 2). Finally, family study revealed that SU172 had inherited the deletion from his mother SU173, in trans of the E47X GJB2 mutation (Figure 2B).
Discussion
A point mutation in the GJB6 gene has been involved in non syndromic deafness (DFNA3) by dominant inhibition of the activity of wild-type GJB6 channels,12 underlining the critical function of GJB6 in normal hearing. Expression studies suggest that GJB2 and GJB6 have a similar regulation in the cochlea, but a possible redundancy of the two encoded proteins seems excluded by the data from Grifa et al.12 This prompted us to suppose that the total absence (homozygous state) of GJB6 protein in itself results in a severe phenotype.
Because we deal with a deletion spanning a large genomic region upstream and including most of the GJB6 gene, several mechanisms could lead to the deafness phenotype. First, removing two doses of GJB6 or removing one dose of GJB6 and one dose of GJB2 result in a reduced number of homomeric or heteromeric connexons that is not sufficient to ensure the correct transport of the potassium ions; then digenic inheritance is the etiology of deafness in the heterozygous cases presented here. Second, the deletion removes the regulatory elements specific for cochlea and/or changes the chromatin conformation, resulting in a functionally null or deficient GJB2 allele. Then deafness results from a GJB2 deficiency alone. Third and last, the profound deafness phenotype is caused by a combination of both mechanisms. Therefore, in addition to the deficit of GJB6, the reduced level of wild type GJB2 may aggravate the phenotype in homozygotes. Then in heterozygotes an allele carrying a GJB2 deafness-causing mutation in trans of a reduced (or null) allele of GJB2, in addition to a deficit of GJB6 causes profound deafness. Expression studies are necessary to evaluate whether this deletion results in a functionally reduced or null GJB2 allele but they remain difficult because of the accessibility of the specific tissue.
There is no doubt that we deal with a similar deletion in the five unrelated patients but we do not know yet whether this deletion is ancestral or recurrent, or both. Finding this mutation in five unrelated patients referred to our clinics prompt us to suppose that this deletion is not rare. Because it requires an appropriate screening it can remain undetected in the heterozygous cases. Although SU1 and SU5 are known to originate respectively from Spain/Italy and Spain/France, the ethnic background of the other patients, who live in France, is unknown. Population studies will be necessary to estimate the occurrence of this deletion and the confirmation of its recessive expression, though the latter is strongly suggested by the data presented here. However, until the exact deletion boundaries are known, large scale screening will remain laborious.
The characterisation of this new GJB6 deletion enabled us to fully determine the molecular basis of deafness in four of the six families initially labelled as ‘heterozygous for a GJB2 mutation.’ Actually, the two remaining families present a distinct deafness etiology. Therefore, at least in specific populations, we emphasise that the screening of this deletion in deaf patients will solve a number of cases heterozygous for one mutation in the GJB2 gene and thereby constitute a significant improvement of the genetic testing and diagnosis.
In conclusion, we describe a new large deletion destroying the GJB6 gene. Considering the importance of GJB6 for normal hearing, we suggest that the homozygous absence of this gene is responsible for severe hearing impairment and therefore DFNB1, as DNFA3, should also include the GJB6 gene. Moreover, this deletion present in trans of a GJB2-deficient allele (35delG or E47X) also leads to profound deafness. That the mechanisms underlying the deafness phenotype reside in a pure digenic effect or in a diminished expression of the normal GJB2 allele, or a combination of both still remain to be proven.
While this manuscript was in review, another group, Lerer et al. (Hum Mutat 2001; 18: 460) has identified a large deletion 5′ of the GJB6 gene in Jewish Ashkenazi families. Though the authors do not show that the deletion lies within GJB6 coding region, their results tend to suggest that the deletion they identified is similar to the deletion presented here.
References
Cohen MM, Gorlin RJ . Hereditary hearing loss and its syndromes in Gorlin RJ, Toriello HV, Cohen MM (eds) Epidemiology, Etiology and Genetic Patterns Oxford: Oxford University Press 1995 pp 9–21
Kelsell DP, Dunlop J, Stevens HP et al. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness Nature 1997 387: 80–83
Zelante L, Gasparini P, Estivill X et al. Connexin26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans Hum Mol Genet 1997 6: 1605–1609
Denoyelle F, Marlin S, Weil D et al. Clinical features of the prevalent form of childhood deafness, DFNB1, due to a connexin-26 gene defect: implications for genetic counselling Lancet 1999 353: 1298–1303
Rabionet R, Gasparini P, Estivill X . Molecular genetics of hearing impairment due to mutations in gap junction genes encoding beta connexins Hum Mutat 2000 16: 190–202
Rabionet R, Zelante L, Lopez-Bigas N et al. Molecular basis of childhood deafness resulting from mutations in the GJB2 (connexin 26) gene Hum Genet 2000 106: 40–44
Denoyelle F, Lina-Granade G, Plauchu H et al. Connexin 26 gene linked to a dominant deafness Nature 1998 393: 319–320
Kumar NM, Gilula NB . The gap junction communication channel Cell 1996 84: 381–388
Stojkovic T, Latour P, Vandenberghe A, Hurtevent JF, Vermersch P . Sensorineural deafness in X-linked Charcot-Marie-Tooth disease with connexin 32 mutation (R142Q) Neurology 1999 52: 1010–1014
Xia JH, Liu CY, Tang BS et al. Mutations in the gene encoding gap junction protein beta-3 associated with autosomal dominant hearing impairment Nat Genet 1998 20: 370–373
Liu XZ, Xia XJ, Xu LR et al. Mutations in connexin31 underlie recessive as well as dominant non-syndromic hearing loss Hum Mol Genet 2000 9: 63–67
Grifa A, Wagner CA, D'Ambrosio L et al. Mutations in GJB6 cause nonsyndromic autosomal dominant deafness at DFNA3 locus Nat Genet 1999 23: 16–18
Maestrini E, Korge BP, Ocana-Sierra J et al. A missense mutation in connexin26, D66H, causes mutilating keratoderma with sensorineural deafness (Vohwinkel's syndrome) in three unrelated families Hum Mol Genet 1999 8: 1237–1243
Richard G, Smith LE, Bailey RA et al. Mutations in the human connexin gene GJB3 cause erythrokeratodermia variabilis Nat Genet 1998 20: 366–369
Lamartine J, Munhoz Essenfelder G, Kibar Z et al. Mutations in GJB6 cause hidrotic ectodermal dysplasia Nat Genet 2000 26: 142–144
Kelley PM Abe S, Askew JW, Smith SD, Usami S, Kimberling WJ . Human connexin 30 (GJB6), a candidate gene for nonsyndromic hearing loss: molecular cloning, tissue-specific expression, and assignment to chromosome 13q12 Genomics 1999 62: 172–176
Lopez-Bigas N, Olive M, Rabionet R et al. Connexin 31 (GJB3) is expressed in the peripheral and auditory nerves and causes neuropathy and hearing impairment Hum Mol Genet 2001 10: 947–952
Xia A, Katori Y, Oshima T, Watanabe K, Kikuchi T, Ikeda K . Expression of connexin 30 in the developing mouse cochlea Brain Res 2001 898: 364–367
Lautermann J, ten Cate WJ, Altenhoff P et al. Expression of the gap-junction connexins 26 and 30 in the rat cochlea Cell Tissue Res 1998 294: 415–420
Lautermann J, Frank HG, Jahnke K, Traub O, Winterhager E . Developmental expression patterns of connexin 26 and -30 in the rat cochlea Dev Genet 1999 25: 306–311
Dahl E, Manthey D, Chen Y et al. Molecular cloning and functional expression of mouse connexin-30, a gap junction gene highly expressed in adult brain and skin J Biol Chem 1996 271: 17903–17910
Beggs AH, Koenig M, Boyce FM, Kunkel LM . Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction Hum Genet 1990 86: 45–48
Acknowledgements
We wish to thank all patients who have participated in this study. We also wish to thank Drs Estivill and Rabionet for initial gift of mutation control DNAs in the GJB2 gene. We are also grateful to Dr Gerd Scherer, Dr Sylvie Tuffery-Giraud and Dr Stéphan Gasca for critical comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pallares-Ruiz, N., Blanchet, P., Mondain, M. et al. A large deletion including most of GJB6 in recessive non syndromic deafness: a digenic effect?. Eur J Hum Genet 10, 72–76 (2002). https://doi.org/10.1038/sj.ejhg.5200762
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200762